The centromere (kentron = center; meros = part) is the region of the mitotic chromosome that participates in chromosomal movement. The mitotic spindle attaches to a specialized structure at the centromere known as the kinetochore. The motor responsible for the movement of the chromosomes toward the spindle poles during mitosis is also located at the centromere. Morphologically, centromeres are distinguished by their appearance at metaphase and anaphase as constrictions in the chromatin and by their heterochromatin staining pattern. A chromosome without a centromere is described as acentric. It does not segregate properly during cell division and is rapidly lost in successive cell cycles.
Centromeres occupy different positions in chromosomes, and they are useful markers. Chromosomes with a single centromere are called monocentric. Chromosomes with centromeres near one end are called acrocentric, those with the centromere visible at or near the middle are metacentric, and those with the centromere truly at the end are telocentric. The monocentric chromosomes found in most metazoan plants and animals almost always have centromeres embedded in segments of heterochromatin. Some organisms, such as plants in the genus Luzula, have holocentric chromosomes with diffuse centromeres. In these chromosomes the spindle attaches to the centromeric heterochromatin that is distributed along the entire length of the chromosome (1). Mammalian tissue culture cells occasionally develop chromosomes with multiple centromeres (2). A chromosome with two centromeres is called dicentric. Dicentric chromosomes are often the result of chromosome breakage followed by fusion. A dicentric chromosome normally breaks at anaphase, when the centromeres are pulled in opposite directions. The holocentric chromosomes found in certain plants have developed as yet unknown mechanisms to avoid this problem.
Centromeres represent highly specialized chromosomal organelles. The simplest types of centromeres are found in the yeast Saccharomyces cerevisiae. The presence of these centromeric sequences, together with the autonomously replicating sequences that serve to direct DNA replication within exogenous plasmid DNAs allow these small minichromosomes to be stably maintained through cell division. The centromeric sequence allows the minichromosomes to be segregated in mitosis and meiosis with accuracy. Maintenance of minichromosomes through cell division also provides a simple assay for the definition of functional centromeric sequences in yeast.
All of the yeast centromeres are functional as small segments of DNA about 1,000 base pairs (1 kbp) or less in length. Nucleosomes are assembled in specific positions on these DNA sequences, and they contain a specialized histone H3 molecule (Cse4p) (16). Within each segment of centromeric DNA are similar nucleotide sequences. In the middle of the centromeric DNA is a 220-bp segment that constitutes the centromeric core, that contains the minimal sequence necessary for centromeric function. Only a single spindle fiber attaches to a yeast centromere (4). If this attachment is to the core, then the nucleoprotein complex assembled on the core also functions as a kinetochore.
Mammalian centromeres are considerably more complex than those in S. cerevisiae. Early experiments on the centromere of the human Y-chromosome established that much more DNA is required for the chromosomal segregation function than in yeast. Over 300 kbp of DNA, including 200 kbp of a-satellite DNA, are required to generate a functional centromere, suggesting the epigenetic—i.e., meta-DNA—mechanisms establish centromere location in cells of metazoa (17), a notion supported by the appearance of neo-centromeres in mammalian cells over nonrepetitive DNA (18). For native centromeres, however, deletion of the a-satellite DNA makes the centromere inactive (5).
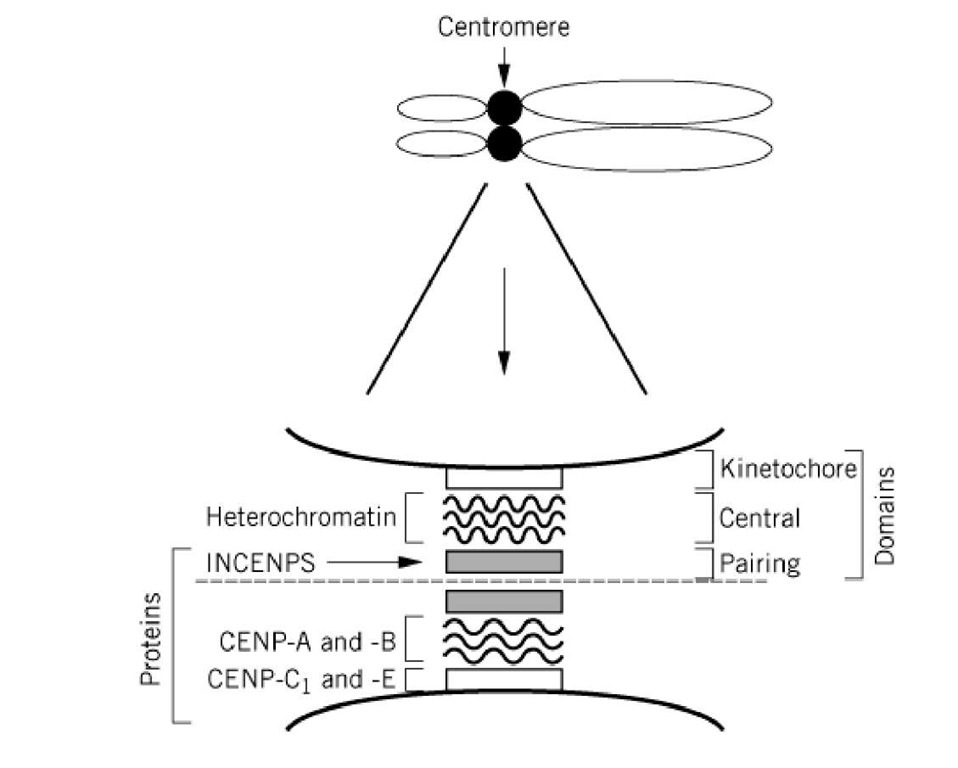
The centromere can be broken up into three distinct structures, the kinetochore and the central and pairing domains (Fig. 1). The protein constituents of the centromere have been identified primarily through the use of autoantibodies from patients with rheumatic disease. These include the inner centromeric proteins (INCENP) and the CENP A, B, and C proteins. Immunological staining reveals that the INCENPs are in the pairing domain. The central domain contains dense chromatin known as constitutive heterochromatin. The kinetochore is anchored to this heterochromatin (Fig. 1). The DNA within this heterochromatin is composed primarily of various families of repetitive DNA (satellite DNA).
Figure 1. An expanded view of the centromere of a mammalian chromosome. The centromere is a bipartite structure. The central axis between the pairing domains is indicated by the dashed line. The relative positions of heterochromatin, the INCENPS, and CENPS are indicated within the kinetochore and the central and pairing domains.
The a-satellite family of DNA sequences (which comprises 5% of the human genome) is probably present at the centromeres of all human chromosomes. The basic repeat 171 bp long, occurs in large arrays up to 3×106 bp long. The a-satellite repeats assemble a specialized chromatin structure that provides many insights into the molecular nature of constitutive heterochromatin. It has been long that nucleosomes are positioned with respect to the nucleotide sequence found in a-satellite DNA so that histone-DNA contacts would begin and end at particular sites (6). An important point is that a single nucleosome is believed to exist on every 171 bp repeat. In modified form, this DNA sequence also provides the foundation for crystallization of the nucleosome more (7). Several interesting specialized chromosomal proteins also associate with a-satellite DNA. A 10-kDa protein, high mobility group protein (HMG-I/Y) binds to a-satellite DNA specifically in vitro (8). The HMG-I/Y protein binds selectively in the minor groove of the double helix, associating with runs of six or more AT base pairs. In addition HMG-I/Y also probably recognizes certain secondary structural features of the DNA molecule. CENP-B specifically recognizes a 17-bp DNA sequence (5′ CTTCGTTGGAA CGGGA 3′) in a subset of a-satellite DNA repeats. The DNA-binding domain of CENP-B is necessary and sufficient for localization to the centromere in vivo (9). CENP-B contains anionic regions rich in aspartic and glutamic acid residues, which are characteristic of many proteins that interact with chromatin. CENP-B is found throughout the centromeric heterochromatin beneath the kinetochore plates (see Kinetochore). Although the exact functions of CENP-B in heterochromatin are not known, it may play a role in the higher order folding of centromeric chromatin through self-assembly mechanisms. Although CENP-B is found at all mammalian centromeres examined, the CENP-B recognition element is not found in all a-satellite DNA (10). It is possible that CENP-B can also be targeted to centromeres through interactions with other protein components.
CENP-A is also specifically associated with centromeric DNA and is especially interestin because it shares homology with core histone H3. Recent sequence analysis has revealed that CENP-A is a very specialized H3 variant (11). Each core histone and CENP-A have two domains, and amino-terminal tail domain that lies on the outside of nucleosomal DNA and a carboxyl-terminal, histone-fold domain that is involved in protein—protein interactions and in wrapping the DNA. CENP-A has a highly divergent amino-terminal tail domain and a relatively conserved carboxy-terminal, histone-fold domain. The targeting of CENP-A to centromeric DNA is directed by the histone-fold domain, and it is very likely that a highly specialized, dedicated chromatin assembly mechanism exists to enable the selective heterodimerization of CENP-A with histone H4 over centromeric DNA (19). Although the exact mechanism by which this is achieved is still unknown. Characterization of a CENP-A homologue in yeast (CSE4, Ref. 3) establishes that this specialized histone is essential for normal chromosomal segregation during mitosis. Genetic evidence suggest that CSE4 heterodimerizes with histone H4 (19). This establishes that specialized nucleosomal structures are present at the centromere, in addition to the presence of positioned nucleosomes. Clearly, the centromere represents a highly differentiated chromosomal domain even at the most fundamental level of chromatin structure.
CENP-C is a large 107-kDa protein that is highly hydrophilic and basic (pI = 9.4) (13). CENP-C is concentrated in a narrow band immediately below the inner kinetochoric plate at the interface between the chromosome and the kinetochore (Fig. 1). CENP-C binds DNA directly and is required for normal kinetochoric assembly. CENP-C is found only at the active centromere of a stable dicentric chromosome, suggesting a direct role in centromeric function (14). Other components of the centromere include CENP-E, which resembles microtubule-binding motor proteins. CENP-E is a 312-kDa polypeptide chain with a tripartite structure consisting of globular domains at the N- and C-termini, separated by a 15,000-residue a-helical domain that is predicted to form coiled-coils. CENE-E colocalizes to the centromere and kinetochore during metaphase, but it is released from the centromere at the onset of anaphase, when it is degraded. CENE-E functions as a kinetochoric motor during the early part of mitosis (15) (See Kinetochore). The proper cohesion between centromeres on sister chromatids prior to the metaphase-anaphase transition in mitosis, and centromere nondisjunction on sister chromatids during meiosis I are essential for proper chromosome behavior. Both are effected by the preferential association of the cohesion complex (120) with centromeres (16, 21); in addition to the cohesins, in metazoa, noncohesion proteins as MEI-S322 also associate with the centromere and mediate kinetochore assembly as well as sister chromatid cohesion during meiosis (22).
The assembly of the centromere and the mechanisms of kinetochoric association with chromatin offer perhaps our best opportunity to understand the construction of a specialized chromosomal domain at the biochemical level.