Barnase is a ribonuclease secreted by the bacterium Bacillus amyloliquefaciens. It is a small protein of 110 amino acid residues with no disulfide bonds or nonpeptide components (see Protein Structure). As such, it was recognized in the early 1960s to be an ideal subject for the then-emerging study of protein folding. Its specific inhibitor, barstar, is produced intracellularly by the same organism and is an equally simple and even smaller protein of only 89 residues. Together they form a two-subunit complex in which the active site of barnase is buried, providing a model system for the study of protein-protein interactions. The system has contributed substantially to our understanding of how proteins fold and interact and has potential for further uses.
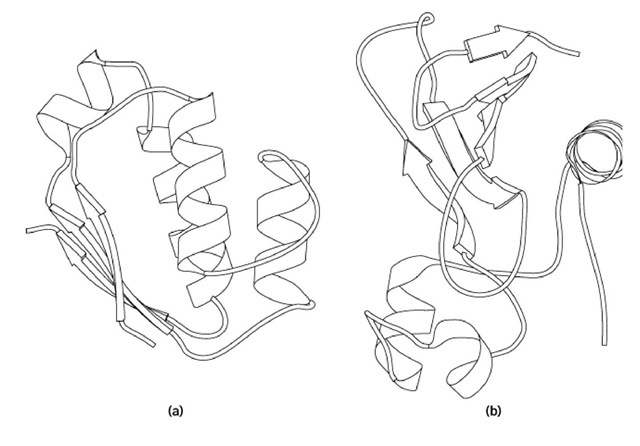
The three-dimensional structures of both proteins and their complex have been determined (Fig. 1) (1). The genes for both proteins have been cloned into plasmid vectors in Escherichia coli and the expressed products obtained in high yield. Expression of barnase alone is lethal, so simultaneous expression of barstar is necessary for production of barnase.
Figure 1. Molscript (1) representation of the structures (a) barstar and (b) barnase in their complex.
The catalytic mechanism of barnase and its relatives is essentially the same as that of the pancreatic ribonuclease family (see Ribonuclease A), with a histidine and a glutamic acid residue acting respectively as proton donor and acceptor, in place of the two histidines of the latter. The barnase active site lies in a broad groove on the side of the b-sheet opposite the major a-helix (see Fig. 1). In the complex, which has a dissociation constant on the order of 10-14 M, barstar completely blocks the active site, with most of its contacts involving its second helix and an adjacent loop. An aspartic acid carbonyl group of barstar occupies the position of the attacked phosphate group of substrate RNA. The rate at which barnase and barstar associate is controlled largely by charged groups, positive on barnase, negative on barstar. Mutation to alanine of any of these charged residues reduces binding but increases the stability of each protein, indicating the evolutionary balance between function and stability.
Both barnase and barstar may be unfolded reversibly by heat or by denaturants such as urea or guanidinium chloride (see Protein unfolding). Both fold and unfold in a highly cooperative two-state manner; that is, only the native and unfolded states are significantly occupied at equilibrium under most conditions. During the kinetics of folding, however, barnase passes through an observable intermediate state. In this it exemplifies the behavior of most larger proteins or protein domains (see Protein folding). Barstar, on the other hand, appears to collapse more directly to the folded state, as do most other smaller proteins. Studies of barnase folding, using kinetic and equilibrium techniques and site-directed mutagenesis, have traced the order in which its parts come together to form the native fold and have estimated the free energies of various interactions in several states that occur during folding.
Barnase is a member of a large family of microbial ribonucleases that share the same basic tertiary structure but have quite divergent sequences; this provides a wider system for approaching the central problem of how the fold is determined by the sequence (see Protein Structure Prediction).
The homologous ribonucleases of the Streptomyces share less than 25% sequence identity with barnase, but the structure of the active-site region is so well conserved that these enzymes are also inhibited by barstar, withdissociation constants as low as![]() consequently, coexpression of barstar permits the high level expression of the cloned genes for these enzymes as well. As the Streptomyces also produce barstar homologues, it seems clear that the extreme structural conservatism of the active site of the ribonucleases is based on the strict requirement of strong binding to the inhibitor.
consequently, coexpression of barstar permits the high level expression of the cloned genes for these enzymes as well. As the Streptomyces also produce barstar homologues, it seems clear that the extreme structural conservatism of the active site of the ribonucleases is based on the strict requirement of strong binding to the inhibitor.
Separate from the continuing application of barnase and barstar as small model proteins to general problems of protein structure and chemistry are more recent uses of their genes that take advantage of the toxic effect of barnase expression in tissues of heterologous organisms. Under control of a specific promoter, expression of the barnase gene can ablate cells under conditions or in tissues where the promoter is turned on. This property is being applied, for example, in studies of development, strategies against viruses, design of a conditionally lethal selective cloning vector, and, most spectacularly, in the development of male-sterile plants. In the last instance, male sterility can be reversed by inclusion of the barstar gene.
There have been several efforts at computer simulation of aspects of barnase folding and unfolding, and more can be expected. The continued application of directed mutagenesis, physical chemistry, and computer modeling to the folding mechanisms of barnase and barstar and their homologues will provide insight into the manner in which the sequences of these two interdependent families of proteins determine their native folds. In vivo use of the barnase gene, with its lethal effect limited to specific conditions or tissues, will be useful to developmental biologists and should have many practical applications in agriculture and possibly in medicine as well.