Molecular genetics provides a powerful new tool for – the conservation and management of marine mammals: the forensic identification of products derived from hunting, strandings, and fisheries bycatch. Such products include soft tissue such as meat, organs, blubber, skin, and blood, as well as teeth, bone, baleen, and hair. Although the species origins of these products may be impossible to determine on the basis of appearance, they contain DNA that can be sequenced and compared to sequences from known specimens. With a complete library of reference sequences, a product of unknown origin can be attributed in most cases to 1 of the (approximately) 80 cetacean species or 40 pinniped species or subspecies. If a comprehensive archive of tissue is maintained as part of a regulated hunt, it is possible to trace the origins of a product to a specific individual.
The molecular methods used in marine mammal forensic genetics have their origins in efforts to isolate and amplify DNA from ancient biological material (Paabo, 1988). With the advent of the polymerase chain reaction (PCR) and “universal” primers (Kocher et al, 1989), DNA can now be recovered from almost any biological source, even products that have been preserved, cooked, or canned. The interpretation of DNA sequences for the identification of species and geographic origins, however, is also dependent on basic research in species-level phvlogenetic relationships and on the genetic structure of populations (Baker and Palumbi, 1995). The application of forensic genetics to marine mammals has, in turn, highlighted inadequacies in the basic taxonomy of cetaceans and pinnipeds and the recognition or definition of subspecies and management units (i.e., “stocks”) below the species level (see later).
The forensic use of molecular genetic methods is of particular interest to the International Whaling Commission (IWC), as it attempts to develop a revised management scheme for the regulation of any future commercial whaling, and to the Convention on Internationa] Trade in Endangered Species of Wild Fauna and Flora (CITES), as it attempts to implement a verifiable system for controlling trade in cetacean products. Progress in the molecular monitoring of commercial markets in whale products is described later as an example of this application. Other applications of forensic genetics to marine mammals include identifying strandings and fisheries bycatch, particularly for poorly described species such as beaked whales (Dalebout et al, 1998; Henshaw et al, 1997), and monitoring of trade in pinniped penises marketed as aphrodisiacs (Malik et al, 1997). These methods are finding similar applications in the conservation and management of other taxa, most notably the hunting and bycatch of turtles (Bowen, 1995) and international trade in caviar (DeSalle and Birstein, 1996).
I. Phylogenetic Identification
Species-level identification of marine mammals has relied primarily on the phylogenetic reconstruction of DNA sequences from the control region or cytochrome b gene of the mitochondria] (mt) genome. In general, mtDNA offers two important advantages over traditional genetic markers such as allozymes. First, because of its maternal inheritance and absence of recombination, the phylogenetic relationship of mtDNA sequences reflects the history of maternal lineages within a population or species. (If hybridization is encountered, nuclear markers are required to identify the paternal species; see later.) Second, all else being equal, the effective population size of mtDNA genomes is one-fourth that of autosomal nuclear genes and its rate of random genetic drift is proportionately greater. This results in more rapid differentiation of mtDNA lineages among populations, compared to nuclear genes, and consequently greater sensitivity in the detection of recent historical demographic or speciation events. The ability to detect population differentiation is also enhanced by the rapid pace of mtDNA evolution, which is generally estimated to be 5 to 10 times faster than nuclear coding DNA in most species of mammals. The control region of the mtDNA does not code for a protein or RNA and, in the absence of these constraints, accumulates mutational substitutions more rapidly than other regions. For this reason, it has become the marker of choice for most studies of the population structure of cetaceans and pinnipeds. The cytochrome b gene, a protein region of the mtDNA, has also been used widely in species-level systematics and, in some cases, for the population structure of marine mammals (Lento et al, 1997). Because of the large number of reference sequences available on public databases such as GenBank and EMBL, both loci have been used for species-level identification of marine mammals.
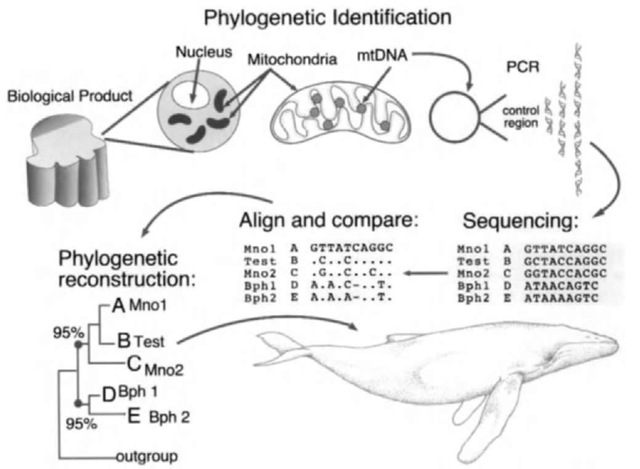
The basic steps involved in the phylogenetic identification of an unknown specimen or product are illustrated in Fig. 1. First, mtDNA is extracted from the product in question, such as a flensed piece of skin and blubber from a commercial market. Second, a fragment of the mtDNA control region is amplified from the product via PCR: a cyclic, in vitro enzymatic reaction that results in the exponential replication of a small targeted fragment of DNA (usually less than 1000 bp). Third, the exact nucleotide sequence of the amplified fragment is determined using a dideoxy-terminator sequencing reaction, followed by electrophoresis through an acrylamide gel. In many laboratories, this step is now automated with a computer-assisted, laser scanner. Fourth, the sequence of the product, now referred to as the “test sequence,” is aligned to and compared with the sequences from reference samples. Finally, the sequence from the product is grouped, by phylogenetic reconstruction, with the most closely related reference sequences. The reconstruction is usually represented as a “tree,” with closely related sequences forming neighboring branches. This allows a hierarchical comparison to establish, first, the suborder and family derivation using a small number of reference sequences from a large number of species. A close relationship or match with a “reference” sequence provides evidence for identification of the species origin of the product. One or more “outgroups” (i.e.. distantly related species) are used to protect against a mis-classification error. The strength of support for an identification or phylogenetic grouping is evaluated by “bootstrap resampling of sequence data. The relative support for a grouping or branch in the tree is shown as the percentage agreement from a large number (>500) of bootstrap simulations.
Figure 1 The basic steps involved in the phylogenetic identification of a whale product using nucleotide sequences amplified by PCR from the mtDNA control region.
As a conservative approach to forensic identification, a species identification should be considered “confirmed” only if the test sequence is “nested within the range of type sequences for a given species. This is necessary because the molecular systematica of some marine mammals, particularly cetaceans, are not fully described (see later). If a test sequence is intermediate between two groups of reference sequences, rather than nested within one or the other, it could be a related species or subspecies not included in the reference database.
When a large set of reference sequences is available from the range of a single species, it is possible to use “intraspecific” variation to evaluate the geographic origin of a sample. In manv cases, the management of marine mammals is based on geographic populations or “stocks” (Dizon et al., 1992). Catch quotas and limits of incidental mortalitv from fisheries bycatch are usually set according to such stock definitions, as well as according to species. Hunting may be allowed in an abundant stock but prohibited in another stock of the same species that is depleted from past exploitation. However, the ability to identify or estimate the stock origins of a specimen or product is determined by the genetic distinctiveness of the recognized stocks, as well as by the comprehensiveness of the reference samples. For some (but not all) species of baleen whales and a few pinnipeds, it is possible to identify the origin of a product to oceanic population and to geographic stock within oceans (see later).
An alternative to the phylogenetic identification of an unknown specimen or product is individual identification by DNA “fingerprinting” or “profiling” using nuclear markers. This is only possible for the control of whale products originating from a regulated hunt or bycatch. As in human forensic genetics, a combination of highly variable nuclear markers can be used to establish individual identity with high probability (or to exclude identity with certainty). The DNA profiles of each individual whale can be stored on a searchable electronic database, forming a “register” of all products intended for the market. If the register is comprehensive (fully diagnostic), a match with a market product would confirm the legality of the product. A product that did not have a match in the register would be illegal. Further genetic investigation would be required to determine the species and geographic origin of illegal products.
II. “Portable Polymerase Chain Reaction”
In some cases, the application of forensic genetics to international conservation problems has involved an additional technical step required by international law. Under the requirements of the CITES, it is illegal to transport native products derived from most marine mammals without both importation and exportation permits from the respective countries of origin and destination. The processing of such permits is a lengthy affair in many countries, and for politically sensitive requests either nation can deny a permit, effectively terminating the research. However, CITES regulations do not apply to “synthetic” DNA created during amplification by PCR, assuming that all “native” product has been removed (Jones, 1994). To comply with these regulations, it has been necessary to assemble a portable PCR laboratory and conduct the initial PCR amplifications from whale products on site in the Japan and South Korea (Baker and Palumbi, 1994). Once the amplified DNA was isolated from the native product, it could be transported internationally for sequencing and final analysis. In this way, the “portable PCR” has become an important tool for international conservation genetics.
III. Monitoring Commercial Markets in Whale, Dolphin, and Porpoise Products
In recognition of historic patterns of overexploitation, the IWC voted in 1982 to impose a global moratorium on commercial whaling. Although the moratorium took effect in 1986, whaling never actually stopped. IWC member nations continue to hunt some species of whales for scientific research or for aboriginal and subsistence use. Whales killed for scientific research can legally be sold to domestic consumers and traded to other member nations of the IWC, thereby sustaining a commercial market for meat, skin, blubber, and other whale products. Small cetaceans are also hunted or taken as fisheries by-catch and sold for consumption in many parts of the world. Although the IWC regulates only hunting of large whales, international trade in all cetaceans is subject to CITES. When some species are protected by an international prohibition against hunting or trade but similar species are not, it is crucial to identify the origin of products that are actually sold in retail markets.
In an effort to monitor the sale and trade of cetaceans products, molecular methods have been used to identify the species and geographic derivation of products sold in two countries with active commercial markets: Japan and the Republic of (South) Korea. Whale meat is widely available in retail markets of both countries despite the international moratorium on commercial whaling. In 1995, high-quality whale meat sold for up to $30/kg in Korea and $460/kg in Japan (Chan et al, 1995). Japan sustains a legal market for whale products by killing several hundred minke whales in the Southern Hemisphere and 100 minke whales in the North Pacific Ocean each year under an exemption to the moratorium for scientific research. South Korea has no program for scientific hunting but reports a substantial fisheries bycatch of cetaceans each year, including up to 128 ininke whales from coastal waters (Mills et al, 1997). It is assumed that products from this unregulated incidental mortality are sold in local markets, but their international trade is prohibited by CITES.
IV. Species, Stock, and Individual Identification
At least 11 independent reports have been published or formally presented to the IWC documenting the molecular genetic identification of nearly 1000 products from commercial markets (Dizon et al, 2000). About 50% of all products were identified as Antarctic minke whale (Balaenoptera bonaeren-sis), the target of scientific hunting by Japan in the Antarctic. About 20% were identified as North Pacific minke whale (Balaenoptera acutorostrata scammoni), the target of Japanese scientific hunting in the North Pacific. The remainder were found to be small cetaceans (including porpoises, family Phocoenidae; and a variety of dolphins, family Delphinidae), beaked whales (family Ziphiidae), sperm whales (family Physeteridae/ Kogiidae), and other protected baleen whales. The latter group included fin (B. physalus), blue (B. musculus) or blue/fin hybrids, Bryde’s (B. edeni), sei (B. borealis), and humpback whales (Megaptera novaeangliae). A few products sold as “whale” were found to be sheep or horse.
Detailed comparisons of mtDNA sequences have provided information on the stock derivation of some commercial products. For example, the North Pacific ininke whale forms at least two stocks with marked genetic differences (Goto and Pastene, 1997): the “J” stock found in the Sea of Japan/East Sea and the “O” stock found in the North Pacific to the east of Japan. Although the “O” stock is subject to legal scientific hunting by Japan and is reported to be relatively abundant, the “J” stock was depleted by commercial hunting before 1986 and is considered a “protection stock” by the IWC. Using molecular methods, market surveys from 1993 to 1999 showed that a large proportion of products from Korea and a substantial proportion from Japan were derived from the protected “J” stock (Baker et al., 2000). Although one source of these products is fisheries bycatch, the possibility of illegal hunting of this stock cannot be excluded.
The detailed analysis of a Japanese market product identified initially as a blue whale shows the importance of maintaining archived tissue from a regulated hunt for verification of trade records. The mtDNA sequence for this product matched closely with the published sequence of a blue/fin hybrid killed during a scientific whaling program by Iceland. Because intDNA is inherited maternally, it cannot, by itself, identify a product as a hybrid. Subsequent comparison of variable nuclear DNA markers from tissue archived during the Icelandic whaling program confirmed that this product was derived from this hybrid individual (Cipriano and Palumbi, 1999).
V. Taxonomic Uncertainties
A final, and critical, consideration in using phylogenetic methods for species identification is the assumption that the taxonomy of the group in question is complete. If this basic biological information is lacking, questions about the adequacy of a reference database and the genetic distinctiveness of recognized species cannot be answered. Surprisingly, taxonomic uncertainties remain a problem among the baleen whales, even though this group includes only a handful of widely distributed, formerly abundant species. For example, two products purchased on Korean markets in 1994 grouped with reference sequences from both the sei and Bryde’s whales but did not group closely with either of these two recognized species (Baker et al, 1996). As a result, it seems likely that diese products originated from an unrecognized species or subspecies of Bryde’s whales. To be fully confident in all identifications, worldwide surveys are needed for nuclear and mtDNA variation among each of the 11 recognized species of baleen whales, particularly the Bryde’s/sei complex and minke whales. Such surveys would provide the basis for an objective evaluation of current distinctions among species, subspecies, morphological forms, and populations or stocks.