Segmentation is the repetition of body units along the anterior-posterior axis and is a fundamental property of all insects; indeed, it is an obvious character of all arthropods. Insect segments are clearly visible as reiterated patterns visible in the exoskel-eton, but repeating patterns are also present in internal structures such as muscles, neurons, and tracheae. Through genetic and molecular approaches in the dipteran fruit fly, Drosophila melanogaster, the mechanisms of segmentation in this insect are now understood in great detail. Recently, interest in the evolution of segmentation has inspired numerous comparative studies of the mechanisms governing segmentation in diverse insect taxa. These experiments indicate that some aspects of the Drosophila mechanisms are conserved in all insects, whereas others have undergone extensive evolutionary changes.
PATTERN O JI SEGMENTS
Segments are most easily visible in the exoskeleton of the adult, where we can see the repeating pattern in the cuticle. The rigid body plates that make up the adult exoskeleton, or sclerites, are separated by membranous intersegmental grooves that lie at the boundary between segments and allow the body to flex. The segmented exoskeleton is also often endowed with reiterated patterns of pigmentation and other elaborations, such as denticles or hairs on the exoskeleton (Figs. 1 and 2 ).
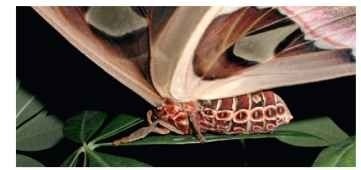
FIGURE 1 The pattern of segments can be easily seen on this Atlas moth (Attacus atlas), made even more evident by the reiterated pigmentation pattern. Note the head, thoracic, and abdominal tagma.
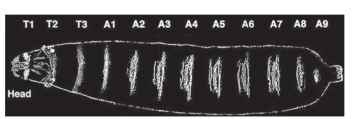
FIGURE 2 The larval segments of Drosophila. The pattern of segments is clearly revealed by the pattern of denticles (hairlike projections) on the ventral surface of the larvae. The segments of the head are involuted inside, and the terminal abdominal segments are not visible on the surface.
Individual insect segments also show various levels of specialization in terms of morphology and function, and are grouped into three primary regions, also known as tagmata: the head, thorax, and abdomen. The head is composed of six segments, the thorax of three, and the abdomen of eleven segments. Segments of the thorax and abdomen are most easy to recognize, whereas the segments of the head and terminal regions of the abdomen are less apparent because they become fused in some insects during development. The head is composed of the antennal, ocular, intercalary, mandibular, maxillary, and labial segments (progressively from anterior to posterior), although some researchers have suggested the existence of a seventh segment at the very anterior of the head. The thorax has been specialized for locomotion and its three segments bear the six legs and the wings. The most posterior abdominal segments (A10 and A11) are fused and reduced in some species, making them difficult to recognize in adults but can usually be detected during embryogenesis.
Although segmentation is most clearly visible in the exoskeleton and in other products of the ectoderm, the pattern of segmentation is also reflected in the arrangement of internal features as well. The musculature, tracheal system, and nervous system are also segmented. The insect central nervous system is laid down in a segmen-tal pattern and contains reiterated ganglia for almost all segments, although some degree of secondary fusion has occurred. Ganglia arise in all abdominal segments during development, but in most insects, the eighth abdominal ganglion is a condensation of the posterior ganglia and innervates the eighth and all succeeding posterior segments. In some insects, a greater level of fusion has occurred. In cyclorra-phous flies and in the bedbug Rhodnius, for example, the entire ventral nerve cord is consolidated into a single mass of nerve tissue.
PARASEGMENTS
The segments that we see in the insect epidermis are so obvious that it is tempting to think that they must represent the fundamental building blocks of the insect body plan. It turns out, however, that the basic unit of segmentation is actually the “parasegment.” Parasegments (para from the Greek, meaning “beside” or “near”) are the same width as traditional segments, but are shifted slightly out of phase from them along the body axis such that a single par-asegment is composed of approximately the posterior one-fourth of one segment and the anterior three-fourths of the next adjacent segment. So if traditional segments and parasegments are both the same width but merely shifted out of phase from each other, why consider parasegments at all?
The evidence for the existence of parasegments and their primacy in segmentation comes from several lines of evidence. First, the expression and mutant phenotypes of several of the homeotic selector genes (described below) seem to affect regions bounded by parasegmental boundaries. Second, the signaling centers established by the segment polarity genes act across parasegment boundaries. Third, during the development of several insect orders, small grooves corresponding to parasegment boundaries can be seen temporarily subdividing the ectoderm before the appearance of the deeper and permanent segmental grooves. Lastly, data from other arthropods suggests that parasegments serve a fundamental role in anterior-posterior patterning in these animals. Analysis of segmentation gene expression in a wide variety of other arthropods also strongly supports the fundamental nature of parasegments throughout the arthropods, and lineage analysis studies performed in some crustaceans has shown that single rows of ectodermal cells divide in stereotyped patterns to give rise to a single parasegment—evidence that the genealogical unit in these crustaceans is the parasegment.
MECHANISMS OF SEGMENTATION IN DROSOPHILA
Studies of insect segmentation through experimental manipulations such as ligation, ablation, centrifugation, and transplantation have a rich history and continue to provide important insights into the mechanisms of segmentation. Beginning about 25 years ago, however, our understanding of these mechanisms was rapidly accelerated by genetic mutant screens in the fruit fly, D. melanogaster. Indeed, these screens uncovered such fundamental principles of biological pattern formation that the geneticists who carried them out, Edward Lewis, Eric Wieschaus, and Christiane Nils slein-Volhard, were awarded the Nobel Prize in Medicine and Physiology for their contributions to the genetic analysis of tagmosis and segmentation in Drosophila. The genetic analysis of segmentation was quickly supplemented with molecular and biochemical studies that have provided detailed knowledge of how segments are generated along the anterior-posterior axis during Drosophila embryogenesis. Because so much is now known about the mechanism regulating segmentation in Drosophila, it has served as a frame of reference for understanding this process in all insects.
Early mutant screens identified many genes that act to establish the pattern of segmentation, and they were grouped into several classes based on their associated mutant phenotypes. These pheno-types indicate that the segmentation genes in Drosophila act in a hierarchical manner to sequentially subdivide the embryo into progressively smaller and smaller units, ultimately establishing the pattern of segments we see in the Drosophila larva (Fig. 3 ).
The Maternal Effect Coordinates Genes
Studies of the maternal effect genes show that the process of segmentation actually begins during oogenesis, when specific messenger RNAs (mRNAs) are localized at either the posterior or anterior end of the developing egg. For example, bicoid mRNA is localized to the anterior end of the egg and forms a gradient of protein in the egg once it has been fertilized (with the highest concentration of bicoid protein at the anterior end). Mothers lacking functional bicoid produce embryos in which the anterior segments are missing. A reciprocal gradient of the nanos protein is also formed, and nanos mutants are missing the more posterior regions of their bodies. The formation of these gradients by simple protein diffusion is possible because the early development of Drosophila is syncytial, with no cell membranes between the nuclei of the early embryo allowing proteins to diffuse across the early embryo. These gradients of information
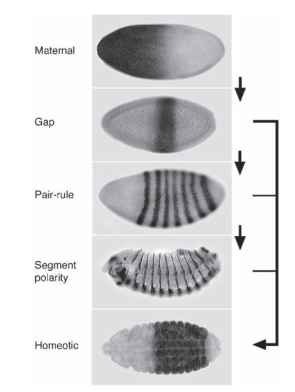
FIGURE 3 The segmentation hierarchy in Drosophila. The hierarchy is composed of the sequential expression of maternal, gap, pair-rule, and segment polarity genes. An example of the expression pattern of a single member of each class is shown here. The home-otic genes act to give regionalization to the segments and are primarily controlled by the gap genes, with some input from the pair-rule and segment polarity genes.
act to control the expression of the various downstream zygotic gap genes, which are the next step in the segmentation hierarchy.
The Gap Genes
The gap genes are so named because mutations in this class of genes cause deletions of several contiguous segments causing a “gap” in the resulting larva. The gap genes read the informational gradients set up by the maternal genes and along with cross-regulatory inputs from other gap genes, become expressed in broad but well-defined domains across the early embryo that roughly correspond to the regions that are deleted in the mutants. All of the gap genes encode transcription factors that act together to regulate expression of the downstream pair-rule genes.
The Pair-Rule Genes
The expression and function of the pair-rule genes reveals the first periodic patterns in the Drosophila embryo. Like the gap genes, the pair-rule class of genes was originally defined through their loss of function phenotypes—in this case, deletions with a two-segment periodicity. Accordingly, most of the pair-rule genes go through a phase of expression consisting of seven stripes—corresponding to a two-segment periodicity—beginning at the syncitial blastoderm stage of the embryo and persisting through cellularization. When the striped pattern for one such pair-rule gene, even-skipped . was first observed, it was thought that the beautiful regularity of the pattern was due to some sort of chemical oscillation that could be modeled with reaction-diffusion equations. Instead, it turns out that stripes are specified individually by the upstream gap and maternal genes acting directly on the DNA regulatory regions that control even-skipped expression. The pair-rule genes encode transcription factors that work together to regulate the final level of the segmentation hierarchy, the segment polarity genes.
The Segment Polarity Genes
The segment polarity genes were also originally identified in genetic screens and named for their mutant phenotypes, which show defects in every segment. These genes are generally expressed in patterns of segmental stripes and include not just transcription factors, but also various receptors, ligands, and enzymes that are used in cell-cell communication, and act to maintain and further refine the pattern of segments that has been elaborated.
The Homeotic Genes
A final category of genes, the homeotic genes, do not act to produce segments, but rather give identity to the segments. Mutations in these genes result in the transformation of one or more segments to the identity of another segment. For example, certain loss of function mutations in proboscipedia cause legs to appear in the place of the adult labial palps. The homeotic genes are primarily regulated by the gap genes, although pair-rule and segment polarity genes also have an important role in defining the precise boundaries of homeotic gene expression. All the homeotic genes encode a family of closely related transcription factors and, in Drosophila, are organized into two complexes on one of the chromosomes. Interestingly, the expression of the home-otic genes along the body axis and their arrangement in the genome are roughly co-linear; homeotic genes expressed in anterior segments of the embryo are situated 3 . in the complex, while genes expressed in the posterior are 5 . in the complex. This co-linear arrangement is highly conserved throughout the bilaterian animals, but the reasons for this chromosomal arrangement are not fully understood.
RELATIONSHIP OF DROSOPHILA SEGMENTATION TO SEGMENTATION IN OTHER INSECTS
Short and Long Germ Segmentation
Although the genetic analysis of segmentation in Drosophila provided an invaluable insight into the mechanisms of pattern formation, earlier manipulative studies in a variety of insects suggested that some aspects of segmentation differ among the various insects and that indeed Drosophila might be somewhat unusual in its mechanisms of segmentation. Drosophila is classified as a long germ insect because various manipulative experiments showed that pattern formation for segments along the entire antero-posterior axis was achieved very rapidly across the entire length of the embryo at the blastoderm stage, without the need for growth. At the molecular level, this is reflected in the nearly simultaneous appearance of the complete complement of pair-rule gene stripes in the Drosophila blastoderm.
Long germ segmentation as typified by Drosophila turns out to be evolutionarily derived. In contrast, the ancestral mode of segmentation is termed short or intermediate germ development and in this case, only the most anterior segments are present at the blastoderm stage (prior to gastrulation), with more posterior segments being added at later stages of development as the embryo elongates. This involves both a dramatic lengthening of the germband as well as the concomitant specification of the remaining segments, in a sequential anterior to posterior progression (Fig. 4). Thus, in short germ segmentation, there is a steep temporal gradient between anterior and posterior patterning along with a phase of secondary posterior growth. It is perhaps surprising that such seemingly differing modes of development should nonetheless converge upon the highly conserved insect body plan. This paradox has led many to investigate how the mechanisms of body segmentation have changed during insect evolution.
Molecular Data in Short Germ Insects
The great depth of our understanding of Drosophila segmentation has led to a recent resurgence of interest in the development of other insects. The embryological differences seen in short and long germ development implies underlying changes at the molecular and genetic level. Homologs of several Drosophila segmentation genes now have been studied in a wide variety of insect taxa and their expression and function show both remarkable levels of evolutionary conservation as well as surprising instances of change.
In all insects examined so far, the homologs of Drosophila segment polarity genes are expressed in stripes as in Drosophila suggesting a conserved role in maintaining parasegment boundaries across diverse insect taxa. But reflecting their short germ mode of segmentation, the stripes appear sequentially in an anterior to posterior progression as the embryos grow.
Moving up the segmentation gene hierarchy, the pair-rule genes show more evolutionary lability. In the red flour beetle, Tribolium casteneum, even-skipped is expressed in a pair-rule like pattern, but again the stripes appear sequentially over time as the embryo elongates. In the milkweed bug, Oncopeltus fasciatus, even-skipped
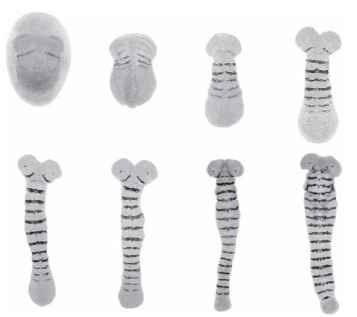
FIGURE 4 Embryos of the intermediate germband insect, Tribolium casteneum. These embryos have been stained for a gene called engrailed, which is expressed as a single stripe in each segment. Six anterior segments have been specified by the end of blastoderm stage (top left) with the remainder appearing sequentially during later stages of development. Compare the sequential appearance of segments in this insect with the progressive subdivision seen during Drosophila segmentation (Fig. 2 ).
bypasses the pair-rule phase of expression entirely and the stripes appear in every segment (both Drosophila and Tribolium even-skipped goes through an early pair-rule pattern followed by a late segmental pattern). In the grasshopper, Schistocerca americana, even-skipped is not even expressed in stripes at all. These differences and differences in expression of other pair-rule homologs suggest that extensive evolutionary alterations have occurred at this step of the segmentation hierarchy, although these changes still result in a conserved output of segment polarity gene expression.
Less work has so far been performed on gap genes in other insects, but the results so far have again shown both conservation and change. Even examples of gap gene conservation are often surprising when viewed in the context of short germ development. For instance, mutations of the gap gene Kruppel in Drosophila delete the thorax and first half of the abdomen. In a short germ insect, since the abdomen is formed after the blastoderm stage, the expectation is that Kruppel would not act in this way in the abdomen. However, when Kruppel function is experimentally depleted in Oncopeltus embryos, this gap phenotype is largely conserved. This unexpected conservation of Kruppel function in a short germ insect implies that in certain cases, the differing modes of segmentation may be underpinned by similar molecular mechanisms.
The earliest steps of pattern formation seem even more labile during insect evolution. For example, it is difficult to imagine how a gradient of bicoid protein can form in grasshopper embryos, given that the entire thorax and abdomen arises as a result of cell proliferation well after the blastoderm stage. Studies suggest that the bicoid gene, a key component of the maternal gradient Drosophila system, only arose within the dipteran lineage. This raises the question as to what gene acts in the stead of bicoid in insects lacking this gene. Recent work in Tribolium has provided one example of how the anterior may be patterned in insects lacking bicoid. In Tribolium, another maternally supplied protein, the product of the orthodenticle homolog OTX1, acts together with Hunchback to carry out a role similar to the Bicoid gradient in Drosophila. Like Drosophila, Tribolium embryos also undergo a prolonged syncitial stage (but since they are short germ, only the anterior is specified), so it is not difficult to imagine how these gradients act to pattern the anterior segments. However, many insects such as Schistocerca lack any form of prolonged syn-citial stage and all segments are patterned within a cellular environment. Thus, the anterior maternal gradients acting in Drosophila and Tribolium are probably not universal among the insects.
It is now clear that extensive modifications have occurred in the segmentation system in different insect lineages, and these changes may reflect adaptive changes in the speed and patterns of oogenesis and early embryogenesis in different insect groups. Nevertheless, the picture that is emerging reveals that the early steps in the Drosophila segmentation hierarchy seem evolutionarily labile with more conservation in the later steps. It is clear that mysteries remain; early insect embryogenesis is remarkably varied, promising still undiscovered mechanisms for generating the segmented insect body plan.
RELATIONSHIP TO SEGMENTATION IN OTHER ANIMAL PHYLA
Insects, while obviously holding an extraordinary fascination, represent only a small part of segmented animal diversity; segmented body plans are also a hallmark of the body plans of annelids and chrodates. Remarkably, many of the genes involved in insect segmentation and regionalization are evolutionarily ancient, and are found today in mutliple phyla. In some cases, conservation is found also at the level of developmental function. In particular, the home-otic genes, which control segment identity, are conserved in both structure and function between flies and vertebrates. On the other hand, homologs of Drosophila segment polarity and pair-rule genes are also well conserved between flies and vertebrates, and usually these proteins still play similar biochemical roles, but in different developmental contexts. For example, the segment polarity gene hedgehog is used in many pattern formation steps in vertebrates, such as patterning the dorsal-ventral axis of the neural tube, but it has no known function in vertebrate segmentation.
Cell-cell signaling based on the Notch pathway is involved in vertebrate segmentation, and recently, work in
