Summary
Genomic sequencing has become a routinely used molecular biology tool in many insect science laboratories. In fact, whole-genome sequences for 22 insects have already been completed, and sequencing of genomes of many more insects is in progress. This information explosion on gene sequences has led to the development of bioin-formatics and several "omics" disciplines, including pro-teomics, transcriptomics, metabolomics, and structural genomics. Considerable progress has already been made by utilizing these technologies to address long-standing problems in many areas of molecular entomology. Attempts at integrating these independent approaches into a comprehensive systems biology view or model are just beginning. In this topic, we provide a brief overview of insect whole-genome sequencing as well as information on 22 insect genomes and recent developments in the fields of insect proteomics, transcriptomics, and structural genomics.
Introduction
Research on insects, especially in the areas of physiology, biochemistry, and molecular biology, has undergone notable transformations during the past two decades. Completion of the sequencing of the first insect genome, the fruit fly Drosophila melanogaster, in 2000 was followed by a flurry of activities aimed at sequencing the genomes of several additional insect species. Indeed, genome sequencing has become a routinely used method in molecular biology laboratories. Initial expectations of genome sequencing were that much could be learned by simply looking at the genetic code. In practice, insects are too complex for a complete understanding based on nucleotide sequences alone, and this has led to the realization that insect genome sequences must be complemented with information on mRNA expression as well as the proteins they encode. This has led to the development of a variety of "omics" technologies, including functional genomics, transcriptomics, proteomics, metabolomics, and others. The vast amount of data generated by these technologies has led to a sudden increase in the field of bioinformatics, a field that focuses on the interpretation of biological data. Developments in the World Wide Web have allowed the distribution of this "omics" data, along with analysis, tools to people all over the world. Integrating these data into a holistic view of all the simultaneous processes occurring within an organism allows complex hypotheses to be developed. Instead of breaking down interactions into smaller, more easily understandable units, scientists are moving towards creating models which encompass the totality of an organism’s molecular, physical, and chemical phenomena. This movement, known as systems biology, focuses on the integration and analysis of all the available data about an entire biological system, and it aims to paint an authentic and comprehensive portrait of biology.
During the past two decades, research on insects has produced large volumes of information on the genome sequences of several model insects. Genome sequencing allows quantificatation of mRNAs and proteins, as well as predictions on protein structure and function. Attempts to integrate this data into systems biology models are currently just beginning. While it is difficult to cover all the developments in these disciplines, we will try to summarize the latest developments in these existing fields. In the first section of this topic, insect genome sequencing and the lessons learned from this will be presented. In the next section, analysis of sequenced genomes using "omics" and high-throughput sequencing technologies will be summarized. In the third part of this topic, an overview of proteomics and structural genomics will be covered. A brief overview of insect systems biology approaches will be presented at the end of this topic.
Figure 1 The whole-genome shotgun sequencing (WGS) method begins with isolation of genomic DNA from nuclei isolated from isogenic lines of insects. The DNA is then sheared and size-selected. The size-selected DNA is then ligated to restriction enzyme adaptors and cloned into plasmid vectors. The plasmid DNA is purified and sequenced. The sequences are assembled using bioinformatics tools.
Genome Sequencing
Almost all insect genomes sequenced to date employed the whole-genome shotgun sequencing (WGS) method (Figure 1). Shotgun genome sequencing begins with isolation of high molecular weight genomic DNA from nuclei isolated from isogenic lines of insects. The genomic DNA is then randomly sheared, end-polished with Bal31 nuclease/ T4 DNA polymerase primers and, finally, the DNA is size-selected. The size-selected, sheared DNA is then ligated to restriction enzyme adaptors such as the BstXladaptors. The genomic fragments are then inserted into restriction enzyme-linearized plasmid vectors. The plasmid DNA is purified (generally by the alkaline lysis plasmid purification method), isolated, sequenced, and assembled using bioinformatics tools. Automated Sanger sequencing technology has been the main sequencing method used during the past two decades. Most genomes sequenced to date employed this technology. Sanger sequencing must be distinguished from next generation sequencing technology, which has entered the marketplace during the past four years and is rapidly changing the approaches used to sequence genomes. Genomes sequenced by NGS technologies will be completed more quickly and at a lower price than those from the first few insect genomes.
Genome Assembly
Genomes and transcriptomes are assembled from shorter reads that vary in size, depending on the sequencing technology used. Contigs are created from these short reads by comparing all reads against each other. If sequence identity and overlap length pass a certain threshold value, they are lumped together into a contig by a program called an assembler. Many assembly programs are available, which differ mainly in the details of their implementation and of the algorithms employed. The most commonly used assembler programs are: The Institute for Genomic Research (TIGR) Assembler; the Phrap assembly program developed at the University of Washington; the Celera Assembler; Arachne, the Broad Institute of MIT assembler; Phusion, an assembly program developed by the Sanger Center; and Atlas, an assembly program developed at the Baylor College of Medicine.
The contigs produced by an assembly program are then ordered and oriented along a chromosome using a variety of additional information. The sizes of the fragments generated by the shotgun process are carefully controlled to establish a link between the sequence-reads generated from the ends of the same fragment. In WGS projects, multiple libraries with varying insert sizes are normally sequenced. Additional markers such as ESTs are also used during the assembly of genome sequences. The ultimate goal of any sequencing project is to determine the sequence of every chromosome in a genome at single base-pair resolution. Most often gaps occur within the genome after assembly is completed. These gaps are filled in through directed sequencing experiments using DNA from a variety of sources, including clones isolated from libraries, direct PCR amplification, and other methods.
Homology Detection
After assembly, sequences representing the genome or transcriptome are analyzed for functional interpretation by comparing them with known homologous sequences. Proteins typically carry out the cellular functions encoded in the genome. Protein coding sequences, in the form of open reading frames (ORFs), must first be distinguished from other sequences or those that encode other types of RNA. Transcriptome analysis is simplified by the fact that the sequenced mRNAs have already been processed for intron removal in the cell. Distinguishing the correct ORF where translation occurs, from 5′ and 3′ untranslated regions, is easily accomplished by a blast search against a protein database, or possibly by selecting the longest ORF. Finding genes in eukaryotic genomes is more complex, and presents a unique set of challenges.
Genomic ORF detection
Detection of ORFs is more complex in eukaryotes than prokaryotes due to the presence of alternate splicing, poorly understood promoter sequences, and the under-representation of protein coding segments compared to the whole genome. If transcriptome data are available, a number of programs exist to map these sequences back to an organism’s genome (Langmead et al., 2009; Clement et al., 2010). This strategy is especially useful when analyzing non-model organisms, or those projects that lack the manpower of worldwide genome sequencing consortiums. In this manner a large number of transcripts can potentially be identified, along with their regulatory and promoter sequences, and information on gene synteny.
De novo gene prediction algorithms often use Hidden Markov Models or other statistical methods to recognize ORFs, which are significantly longer than might be expected by chance. These algorithms also search for sequences containing start and stop codons, polyA tails, promoter sequences, and other characteristics indicative of protein coding segments (Burge and Karlin, 1997). De novo gene discovery is partially dependent on the organism used, since compositional differences such as GC content and codon frequency introduce bias, which must be considered for each organism. Artificial intelligence algorithms can be trained to recognize these differences when a sufficient number of protein coding sequences are available. These may originate from transcriptome sequencing, or more traditional approaches such as PCR amplification and Sanger sequencing of mRNAs. Based on a small sample proportion of known genes, artificial intelligence programs can learn the codon bias and splice sites, for example, and extrapolate these findings to the rest of the genome. However, this process is often inaccurate (Korf, 2004).
Comparative genomics is the process of comparing newly sequenced genomes to more well-curated reference genomes. Two highly related species will likely have well conserved protein coding sequences with similar order along a chromosome. The contigs or scaffolds from a newly assembled genome can be mapped to the reference, or the shorter reads can be mapped and assembled in a hybrid approach. Programs that perform this task may often be used to map transcriptome data to a genome, since the two approaches are mechanistically similar.
Transcriptome gene annotation By definition, mRNA represents protein coding sequences, and finding the correct ORF requires only a blast search. However, ribosomal RNA (rRNA) may represent more than 99% of cellular RNA content. The presence of rRNA may be detrimental to the assembly process because stretches of mRNA may overlap, and thus cause erroneously assembled RNA amalgams. Strategies to reduce the amount of sequenced rRNA include mRNA purification and rRNA removal. Oligo (dt) based strategies, such as the Promega PolyATract mRNA isolation kit, use oligo (dt) sequences which bind to the poly A tail of mRNA. The poly T tract is linked to a purification tag, such as biotin, which binds to streptavidin-coated magnetic beads. The beads can be captured, allowing the non-poly adenylated RNA to be washed away. The Invitrogen Ribominus kit uses a similar principle, except oligo sequences complementary to conserved portions of rRNA allow it to be subtracted from total RNA.
During RNA amplification, oligo (dt) primers may be used to increase the proportion of mRNA to total RNA. This process may introduce bias near the 3′ side of mRNA, and thus protocols have been developed to normalize the representation of 5′, 3′, and middle segments of mRNA (Meyer et al, 2009). If the rRNA sequence has already been determined, many assembly programs can be supplied a filter file of rRNA and other detrimental contaminant sequences, such as common vectors, which will be excluded from the assembly process.
Homology detection Annotation is the step of linking sequences with their functional relevance. Since protein homology is the best predictor of function, the NCBI blastx algorithm (Altschul et al. , 1990) is a good place to start in predicting homology and thus function. The blastx algorithm translates sequences in all six possible reading frames and compares them against a database of protein sequences.
For less technically inclined users, the blastx algorithm may be most easily implemented in Windows-based programs such as Blast2GO (Conesa et al., 2005; Conesa and Gotz, 2008; http://www.blast2go.org/). Blast2GO offers a comprehensive suite of tools for blasting and advanced functional annotation. However, relying on the NCBI server to perform blast steps often introduces a substantial bottleneck between the server and querying computer. Local blast searches, performed by the end user’s computer(s), may significantly reduce annotation time. The blast program suite and associated databases may be downloaded for local blast searches (ftp://ftp.ncbi.nlm. nih.gov/blast/executables/blast+/LATEST/). The NCBI non-redundant protein database is quite large and time consuming to search. Meyer et al. (2009) advocate a local approach where sequences are first queried against the smaller, better curated swiss-prot database, and then sequences with no match are blasted against the NR protein database (Meyer et al., 2009). Faster algorithms such as AB-Blast (previously known as WU-Blast) may also speed up the blasting process. After a blastx search, sequences may be compared to other nucleotide sequences (blastn), or translated and compared to a translated sequence to help identify unigenes, or unique sequences. However, blastx is the first choice, since the amino acid sequence is more conserved than the nucleotide sequence. This step will also yield the correct open reading frame of a sequence. In some cases, homologous relationships may be discovered using blastn and tblastn where blastx did not. The statistically significant expectation value, or the probability that two sequences are related by chance (also called an e value) is an important consideration in blasting, because setting an e value too low may create false relationships, while setting an e value too high may exclude real ones. As sequence length increases, the probability of finding significant blast hits also increases. In practice, blasting at a low e value and small sequence overlap length initially, and then filtering the results based on the distribution of hits obtained, may be beneficial.
Gene Ontology Annotation
Gene Ontology (GO) provides a structured and controlled vocabulary to describe cellular phenomena in terms of biological processes, molecular function, and subcellular localization. These terms do not directly describe the gene or protein; on the contrary they describe phenomena, and if there is sufficient evidence that the product of a gene, a protein, is involved in this phenomenon, then the probability increases that a paralogous protein is involved (Ashburner et al., 2000).
For example, GO analysis for the Drosophila melano-gaster protein Tango molecular functions indicates that it is a transcription factor which heterodimerizes with other proteins and binds to specific DNA elements and recruits RNA polymerase. The evidence shows what types of experiments or analyses were performed to determine the function. The GO evidence codes can be inferred experimentally from experiments, assays, mutant phenotypes, genetic interactions or expression patterns, as well as computationally from sequence, sequence model, and sequence or structural similarity. The biological processes information shows that Tango is involved in brain, organ, muscle, and neuron development. The cellular components information indicates that Tango’s subcellular localization is primarily nuclear. Gene Ontology annotation programs often allow the user to set evidence code weights manually. For example, evidence inferred from direct experiments may provide more confidence than evidence inferred from computational analysis which has been manually curated. Uncurated computational evidence may contain the least confidence level. Tango and its human paralog, the Aryl Hydrocarbon Receptor Nuclear Translocator (ARNT), are both well-studied proteins. However, when using the Tribolium castaneum sequence, for example, a good GO mapping algorithm must decide how to report the more relevant information on TANGO without losing pertinent information about the better studied ARNT.
Gene ontology mapping is great when a well-studied parologous protein is available and the blast e value is low enough to provide statistical confidence in the evolutionary relatedness and conservation of function between two proteins. In our example, the user now has a wealth of information about the T. castaneum Tango function, and can design primers for qRTPCR, RNAi, protein expression, or link function to the mRNAs which may have changed between two treatment groups in a tran-scriptome expression survey such as microarray analysis.
Enzyme codes are a numerical classification for reactions that are catalyzed by enzymes, given by the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB) in consultation with the IUPAC-IUBMB Joint Commission on Biochemical Nomenclature (JCBN). Enzyme codes can be inferred from GO relationships.
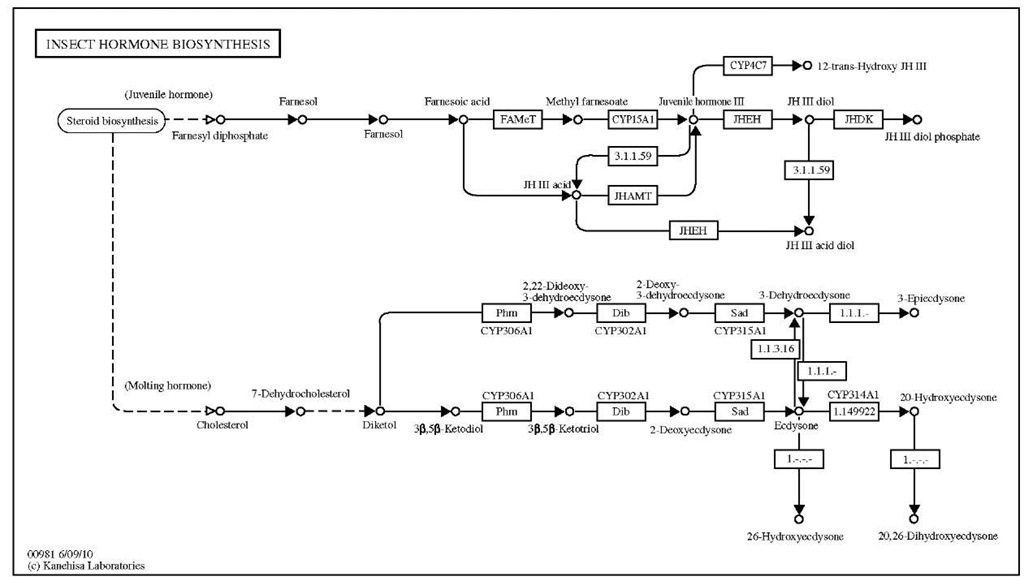
KEGG is an integrated database resource consisting of systems, genomic, and chemical information (Kanehisa and Goto, 2000; Kanehisa et al, 2006). The KEGG pathway database consists of hand-drawn maps for cell signaling and communication, ligand receptor interactions, and metabolic pathways gathered from the literature. Figure 2 shows the pathway for D. melanogaster hormone biosynthesis annotated in KEGG. The information in this database could help in interpretation of data from genome analysis employing "omics" methods.
Conserved Domains and Localization Signal Recognition
Conserved domains often act as modular functional units and can be useful in predicting a protein’s function.
Domain detection algorithms do not require an absolute paralog to predict function, but often use multiple sequence alignments and Hidden Markov Models based on a number of homologous proteins that share common domains. Examples include SMART (Schultz et al., 1998), PFAM (Finn et al, 2010), and the NCBI Conserved Domain Database (CDD) (Marchler-Bauer et al., 2002). Some databases, such as SCOP (Lo Conte et al., 2002), CATH (Martin et al, 1998), and DALI (Holm and Rosenstrom, 2010), focus on structural relationships and evolution. These databases group and classify protein folds based on their structural and evolutionary related-ness. Domain recognition programs have strengths and weaknesses depending on their focus, algorithm implementation, and the database used. Interproscan (Zdobnov and Apweiler, 2001) is a direct or indirect gateway to the majority of these programs and the information they can reveal. Interproscan may be accessed on the web, or through the Blast2GO program suite. Other programs accessed via Interproscan allow the identification of localization signals (i.e., nuclear localization signals), trans-membrane spanning domains, sites for post-translational modifications, sequence repeats, intrinsically disordered regions, and many more.
Fisher’s Exact Test
Perturbations in the expression levels between two treatment groups of gene products involved in GO phenomena or KEGG signaling, or which belong to domain/protein families, can indicate the physiologic effects of the treatment and the mechanisms that are ultimately responsible for changes in phenotypes.
Figure 2 The pathway for D. melanogaster hormone biosynthesis annotated.
mRNA expression changes must be tested for statistical significance to ensure that changes between treatments are not the result of sampling a variable population. Fisher’s Exact Test calculates a p-value which corresponds to the probability that functional groups are over-represented by chance. A low p-value might indicate that the over-represented functional groups share some regulatory mechanism which was perturbed by treatment.