General structure
There are many complex and detailed three-dimensional structures of the CYPs available online which are worth viewing. However, there is perhaps a simpler way to help you visualize some idea of CYP flexibility and structure at the same time. If you place a small coin a little off centre of the palm of your hand towards your first finger, you could imagine that this is the haem iron catalytic centre, the active site of the CYP. If your hand is partially clenched, but not forming a tight fist around the coin, you might see how there are various flexible ‘access channels’ or entrances where a ‘substrate’ can enter and a ‘product’ leave. You can also see how flexible your fingers are in assisting the ‘binding’ of various substrates of different shapes to the less mobile ‘catalytic centre’ of the palm of your hand. In fact, to some extent, we can superimpose the actual generic structural features of real CYP isoforms onto this basic ‘CYP hand’ analogy. The haem – iron active site ‘palm and coin’ is set inside what is sometimes termed the CYP protein ‘ fold’ (your hand). This consists of many coils of protein, known as α helices. The whole enzyme structure is usually anchored (the wrist) in the membrane of the smooth endoplasmic reticulum (SER) by an N-terminal α helix, rather like the legs of an oil rig reaching down to the seabed. The structural features of a CYP are often referred to as distal (far from) or proximal (close to) the haem-iron. Hence, the substrate enters the distal area of the isoform (wrist/palm edge/fingers), whilst the REDOX partners which provide the electrons to operate the enzyme, are proximal to the haem iron (near the thumb and first finger).
Haem moiety
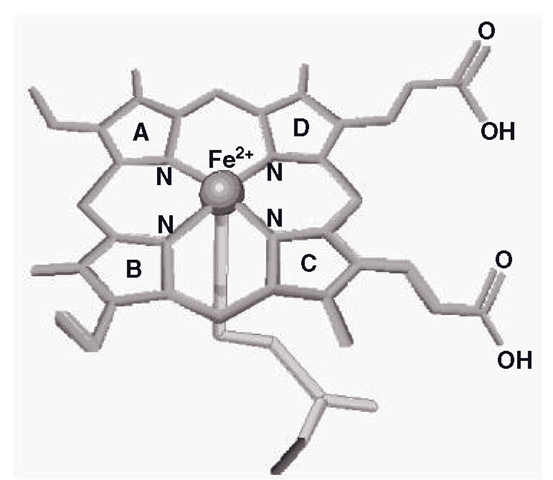
Among the major core α helix sub-structures of CYPs, the backbone of these enzymes is known as the ‘I’ helix, which has a kink in it which locates an area called the ‘cys pocket’, which in turn holds the haem-iron active site in place. This could be the ‘palm and coin’ of the CYP hand analogy. CYPs such as 3A4 and 2C9 have some flexibility in the movement of the haem, but in most CYPs this is a relatively rigid part of the protein’s structure. The haem structure is also known as ferriprotoporphyrin-9 (F-9; Figure 3.2). The F-9 is the highly specialized lattice structure that supports a CYP iron molecule, which is the core of the enzyme, which catalyzes the oxidation of the substrate. This feature is basically the same for all CYP enzymes; indeed, F-9 is a convenient way of positioning and maintaining iron in several other enzymes, such as haemoglobin, myoglobin and catalase. The iron is normally secured by attachment to five other molecules; in the horizontal plane, four of them are pyrrole nitrogens, whilst the fifth group, a sulphur atom from a cysteine amino acid residue holds the iron in a vertical plane. This is known as the ‘pentacoordinate’ (five-position) state and could be described as the ‘resting’ position, prior to interaction with any other ligand (Figure 3.2).
Figure 3.2 Main structural features of ferriprotoporphyrin-9, showing the iron anchored in five positions (pentacoordinate form). The cysteinyl sulphur holds the iron from below
The pentacoordinate state appears to show iron bound tightly to the sulphur and below the level of the nitrogens. When the iron binds another ligand, it is termed ‘hexacoordinate’ and iron appears to move ‘upwards’ and draws level with the nitrogens to bind a water molecule which is hydrogen bonded to a threonine amino acid residue that is located just above the iron, which is linked with proton movements during operation of the enzyme. The F-9 is held in place by hydrogen bonding and a number of amino acid residues, particularly an argenine residue, which may also stabilize the F-9 molecule. As mentioned previously, the iron is crucial to the catalytic function of CYP enzymes, and the process whereby they oxidize their substrates requires a supply of electrons, which is sourced by the dual ‘fuel pumps’ of the system, POR and cytochrome b5. These REDOX partners are sited extremely close to a relatively flat area of the proximal side of the CYP and we will look at them in more detail later on, in section 3.4.6.
CYP flexible regions
Running across and over the ‘I’ helix/haem active site, is a series of helices which form a cover or lid on the active site. These helices are usually described as the F and G ‘domain’, This domain consists of an F helix, and F/G loop structure and a G helix. A B/C helix loop is also part of the cover of the active site. Human CYPs contain extra F and G helices (usually termed F and G’) which give them added flexibility in uncovering the active site enough to accommodate large molecules. To follow the CYP hand analogy, these flexible regions to the structures, the various F/G helices, their loops and the B/C helix loop, could be regarded as the ‘fingers’ which are normally partially clenched, but can open out to form an access pathway to accommodate large substrates. As the human hand can grasp a pin or a beach ball, the small substrate metyrapone binds to CYP3A4 without any visible movement in the ‘fingers’, whilst erythromycin requires them to stretch out widely to allow binding to the active site. Indeed, CYP3A4 increases its active site area by 80 per cent to accommodate erythromycin. To try to see how the whole CYP isoform is oriented in the SER membrane, then you could imagine the lipophilic substrates diffusing through the membrane and entering the isoform through the access path, which includes the highly lipophilic opening ‘fingers’. In any CYP, the access path is generally defined as the widest, shortest and usually the most lipophilic route to the haem iron active site. The active site is supplied with electrons through another access channel from the other side of the CYP by POR and cytochrome b5. As mentioned above, in the CYP hand analogy, this area could be visualized as located between the thumb and first finger. These REDOX partners are also embedded in the SER membrane right next to the CYP. Finally, there is also what is often termed an egress channel (between the ‘fingers’), which is routed away from the SER membrane into either the lumen of the SER or the cytosol, where the more hydrophilic product will naturally exit the isoform, as the other paths are so lipophilic they effectively repel the product.
Substrate binding in CYPs
The term ‘active site’ of an enzyme usually means the area where structural changes in the substrate are catalyzed with the help of various co-factors. This term can encompass a binding area which locates and holds the substrate in such an orientation that the appropriate moiety of the molecule is presented to the structures on the enzyme that catalyze the reactions the enzyme is intended to accelerate. In many enzymes, the dimensions and properties of the active and binding sites are quite well defined and mapped in detail. With acetylcholinesterase for example, the anionic site is mainly responsible for attracting and locating the substrate acetylcholine and the esteratic site is intended to catalyze the hydrolysis of the substrate. This is not the case in CYPs, as crystallographic studies have shown that what constitutes the ‘active and binding sites’ of a CYP can be a very broad area indeed. To date, it has been generalized that CYP3A4, CYP2C8 and CYP2C9 have very large active sites, whilst that of CYP2D6 is intermediate and CYP2A6’s site is quite small. However, with the larger sited CYPs like CYP3A4 and CYP2C9, they can still bind very small substrates alongside the giant ones. If we return to the ‘CYP hand’ analogy, if you grasp an object like a door key, which is similar in size to the coin at the ‘catalytic centre ’ ’ the ’ binding site ’ is a relatively small area on the palm and little hand/finger movement is required to grasp it. If you grasp a dinner plate, you can easily hold it in such a way that the edge contacts the coin, although all your fingers and thumb are now required to articulate to grasp the plate and the CYP ‘binding site’ is pretty much your whole hand. With real CYPs, they undergo similar huge changes in movement and binding area to accommodate substrates of differing sizes like the contrasting agents metyrapone and erythromycin mentioned earlier. Essentially, what constitutes the ‘binding site’ of any given CYP is very difficult to define. Examination of crystallized CYPs bound to different substrates have shown that CYPs do contain small-intermediate hydrophobic pockets, as well as a capability of the rest of the F and G helices to act as ‘extending and enclosing fingers’ to bind larger substrates. What is usually described as the hydrophobic pocket in a CYP comprises many amino acid residues that can bind a molecule by a number of means, including weak van der Waals ‘ forces, hydrogen bonding, as well as other interactions between electron orbitals of phenyl groups, such as ‘ pi -pi bond stacking ‘ , This provides a grip on the substrate in a number of places in the molecule, preventing excessive movement. Interestingly, when not binding substrates, CYP active site areas are full of water molecules, which are displaced upon substrate binding.
In effect, crystallographic studies have shown that the type of hydrophobic amino acid residues seen in the smaller, Internal’ CYP hydrophobic pockets are also found on the ‘fingers’ of CYPs, such as CYP3A4. These are the F and F’, G and G’ helices (among others) and they are capable of binding a hydrophobic molecule by using the same pi-pi bond stacking and van der Waals forces. This effect is borne out by observations of the binding of progesterone, which appears to be held between the ‘fingers’ of CYP3A4, rather than in the ‘palm’, Technically, progesterone actually appears to be stuck to the outside of the enzyme. So it is not unusual for many CYPs, that large swathes of the interior and exterior of the isoforms are available for substrate binding.
It is also apparent from crystallographic studies, that there is much more about CYP binding to discover. Studies have shown that substrate binding is not always ‘productive’, in that the substrate may not be bound near enough to the active site to actually be oxidized (such as a published crystallographic models of CYP2C9 and CYP3A4 with S-warfarin and testosterone binding respectively). Hence, binding may occur in non-productive and productive stages, involving internal rearrangement of the isoform, or may involve simultaneous binding of other substrates. There is also evidence that other conformational changes in the F and G helices occur in contact with an oxidized product molecule, which effectively facilitates its passage out of the CYP.
CYPs: summary of structure and function
CYPs such as CYP3A4 and CYP2C9 managed to combine binding features which at first appear to be contradictory. These and other CYPs can not only bind a range of sizes of substrates, but their sophisticated binding control processes also enable them to even distinguish between left and right-handed versions of the same molecule. This is achieved because we have seen that their structures maintain some areas of relative rigidity and others of constant fluidity, like the relatively rigid palm and flexible fingers of the human hand. Rigidity in the haem promotes oxidation of the substrates, whilst the proximal side of the CYP is sufficiently flexible to link with the REDOX partners, but rigid enough to conduct electrons from the partners to the haem iron. The highly flexible and lipophilic distal side buried in the membrane allows the entrance of virtually any size and shape of molecule and can even facilitate simultaneously the temporary binding of the main body of the molecule during orientation of the target moiety to the haem. In essence, CYPs manage paradoxically to be both broad and highly specific in terms of substrates by ensuring that flexibility in one area does not impair rigidity in the others and vice-versa.
In modern cars, when you turn the key in the ignition, fractionally before the starter motor turns the engine, the electronic management system engages the fuel pump to raise the fuel system to the correct pressure for start up. Similarly, when a substrate binds a CYP, it causes the CYP to become ‘coupled’ with one or other of its REDOX partners which then supply it with sufficient reducing power to oxidize the substrate. The next two sections describe these vital components of CYP function, the REDOX partners.
CYP REDOX partners (i) P450 oxidoreductase (POR)
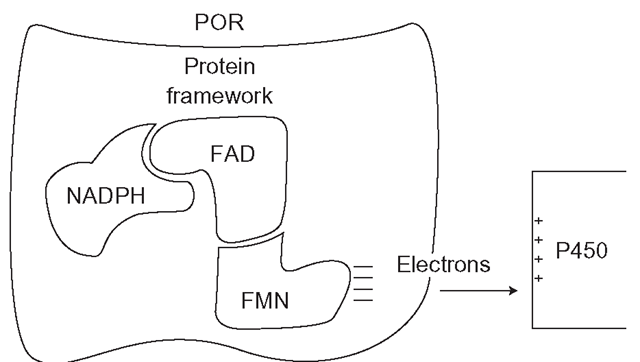
P450 oxidoreductase (POR) is an NADPH reductase that is a separate entity from mammalian CYPs but it is indispensible to them (Figure 3.3). NADPH reductases in general are found in most tissues, but they are particularly common in the liver. POR is essential to life, as removal of the gene from animal embryos is lethal. There is a rare human condition known as Antley-Bixler Syndrome (ABS) which is linked with POR mutations and in severe form results in major structural malformations which are associated with disordered steroid metabolism. Perhaps logically, the expression of POR is mainly under the control of the same nuclear receptors that control CYP expression, such as HNF4a and CAR.The reductase is a flavoprotein complex, which consists of a large vaguely butterfly-shaped protein ‘framework’, which locates and binds two equal components, FAD (flavin adenine dinucleotide, an electron carrier) and FMN (flavin mononucleotide). The structure is unusual in enzymology, as FAD and FMN do not usually function together in the same enzyme, except in the various nitric oxide synthetases. Although in tissues, NADH (used in oxidative metabolic reactions) can be plentiful, FAD has evolved to discriminate strongly in favour of NADPH, which fuels reductive reactions. NADPH is formed by the consumption of glucose by the pentose phosphate pathway in the cytoplasm. This oxidative system, which can consume up to 30 per cent of the glucose in the liver, produces NADPH to power all reductive reactions related to CYPs, fatty acid and steroid synthesis, as well as the maintenance of the major cellular protectant thiol, glutathione.
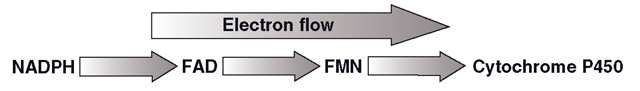
POR will not pass electrons to a CYP unless it is embedded in the SER membrane very close to the CYP. The enzyme complex operates as follows: FAD is reduced by NADPH, which is then released as NADP+ (Figure 3.3). FAD then carries two electrons as FADH2 that it passes on to FMN, forming FMNH2, which in turn passes its two electrons to the CYP. In the presence of high substrate concentrations, POR is required to provide a ‘current’ of electrons to sustain continuous CYP catalytic activity, rather like a machine tool would need electricity in a factory (Figure 3.4). Interestingly, POR is probably not the only source of electrons for the CYP catalytic cycle, which is discussed in the next section.
In the lung, POR is toxicologically relevant, as it mediates the metabolism of the herbicide paraquat, which occasionally features in accidental and suicidal poisonings. The reduction of paraquat by POR leads to a futile cycle that generates vast amounts of oxidant species, which destroy the non-ciliated ’ Clara’ cells of the lung leading to subsequent death several days later. To date there is still no known antidote to paraquat poisoning and it is an agonizing and drawn-out method of suicide. Oxidoreductases are also implicated in the reduction of nitroaromatic amines to carcinogens.
Figure 3.3 Position of CYP reductase in relation to CYP enzyme and the direction of flow of electrons necessary for CYP catalysis
Figure 3.4 Direction of electron flow in P450 oxidoreductase (POR) supply of reducing power to CYP – mediated metabolic processes
CYP REDOX partners (ii) Cytochrome b5
Cytochromes b5 are electron transport haemoproteins which strongly resemble the active sites of CYP P450’s, in that they also are built around a central F-9 haem group. These proteins are ubiquitous in nature, where they convert plentiful cellular supplies of NADH to NAD+, so building up proton gradients which in turn stimulate the flow of electrons. There are numerous forms and structures of cytochrome b5’ a soluble form is found in erythrocytes, where it is known as NADH-dependent methaemoglobin reductase, or sometimes NADH diaphorase. This version of cytochrome b5 converts methaemoglobin,which is formed normally in small amounts and cannot carry oxygen, back into haemoglobin. Interestingly, those with a rare genetic absence of NADH diapho-rase spend their whole lives with blue cyanotic skin. Other forms of the enzyme are lipophilic and membrane bound, such as a variant which is found in the outer membrane of hepatic mitochondria (known as OM cytochrome b5) which is functionally very different to microsomal cytochrome b5 , which is of greatest interest in drug metabolism.
Our understanding of the role of microsomal cytochrome b5 in the operation of CYPs is far from complete but has advanced considerably over the last few years. Microsomal cytochrome b5 is anchored by a helix which penetrates deep into the SER membrane, but its haem structure is cytosolic (it stands clear of the SER membrane) and it is physically linked with its CYP isoform and it is also closely associated with POR. Until relatively recently, it was widely assumed that the flow of electrons needed to ‘power’ CYPs was entirely dependent on POR, which required an adequate supply of NADPH to operate. However, it appears that cytochrome b5 also has a complex but essential role in CYP function. The standard CYP catalytic cycle (see section 3.7 and Figure 3.5) requires two electrons to undergo a complete ‘turn’. The second electron is the ‘rate limiting step’, in that the speed of the CYP operation depends on how rapidly this electron can be supplied. It is now clear that cytochrome b5 can supply this electron just as quickly and possibly even quicker than POR/NADPH. This suggests that CYPs can source their electron flow indirectly from NADH as well as NADPH.
Although work has been carried out using antibodies to inactivate cytochrome b5 which has shown it is likely to be essential for CYP activity, a key study in 2008 actually deleted the expression of hepatic microsomal cytochrome b5 expression in mice. This work showed firstly that the mice suffered no ill-effects and developed normally, which suggests that other enzyme systems can carry out the main functions of cytochrome b5 in cellular housekeeping. What was particularly interesting about this study was that both -n vitro and in vivo, drug clearance by CYPs was greatly reduced by up to 90 per cent of normal with some drugs. This means that in the mouse, not only do most CYPs rely on cytochrome b5 to be part of the process of electron supply, but also the utilization of NADPH and the efficient function of POR is also conditional on the presence of functional cytochrome b5. If these processes occur in man, then it is likely that the two REDOX partners are not only interdependent, but their individual contributions to the supply of electrons may be exquisitely modulated. Indeed, there is some evidence in bacterial CYPs that REDOX activity is coupled to substrate binding and cytochrome b5 binding can even change the structure of the main CYP, changing dimensions of access channels and even binding characteristics. It is likely that this process operates both ways as CYPs and REDOX partners couple and uncouple according to substrate binding and processing.
The full complexity and flexibility of the modulation of CYP substrate binding and the resultant internal enzymatic conformational changes remain to be uncovered. It is likely that the CYP’s response to a particular substrate is effectively ‘customized’ in terms of binding, electron supply and catalytic activity. In some ways CYPs can be regarded as highly sophisticated pumps, which draw up lipophilic agents from the SER membrane ‘conveyor belt’ and expel them as processed and more hydrophilic products into the lumen, thousands of times per second.