Abstract
Cardiomyopathies are included in an heterogeneous group of diseases, characterized by different signs and symptoms, natural history, clinical outcome, and different pattern of inheritance. The genetics of cardiomyopathies has born in 1989 with a single gene theory (one gene=one disese), but the complexity and wide heterogeneity of the disease has moved toward a different direction (one gene=many diseases, or genocopies). Elucidation of the molecular basis of cardiomyopathies has led to a categorization of the phenotypes according to their genetic etiology. The American Hearth Association and the European Society of Cardiology have recently proposed a different scheme of classification based on a distinction between primary (genetic, mixed, non genetic types) and secondary cardiomyopathies, or between the familial and non familial types, respectively. The possibility of a different approach of intervention (i.e. enzyme replacement therapy in metabolic cardiomyopathies) underlies the need to make an early and precise etiologic diagnosis.
Introduction
Cardiomyopathies are diseases of heart muscle. They represent an important and heterogeneous group of diseases. The awareness of cardiomyopathies in both the public and medical communities historically has been impaired by persistent confusion surrounding definitions and nomenclature.For more than 30 years, the term "cardiomyopathies" has been used to describe disorders of the heart with particular morphological and physiological characteristics.
Old Classifications
In 1980, the World Health Organization (WHO) in defined cardiomyopathies as "heart muscle diseases of unknown cause," to distinguish cardiomyopathy (including: Hypertrophic cardiomyopathy, Dilated cardiomyopathy, and Restrictive cardiomyopathy) from cardiac dysfunction due to known diseases such as hypertension, ischemic heart disease, or valvular disease. Heart muscle disorders of known aetiology (eg, ischemic or hypertensive cardiomyopathy) were classified as secondary diseases [1].
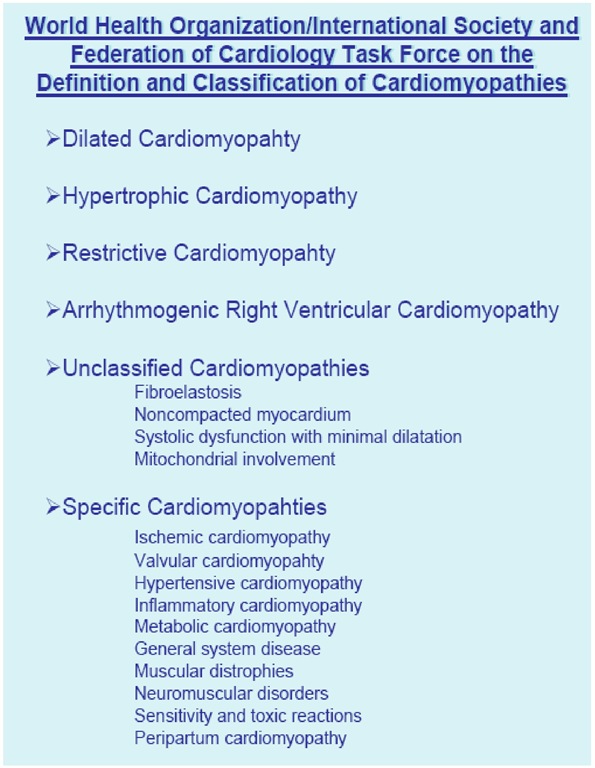
In1995, the WHO/International Society and Federation of Cardiology (ISFC) Task Force on the Definition and Classification of the Cardiomyopathies expanded the classification to include all diseases affecting heart muscle and to take into consideration etiology as well as the dominant pathophysiology. Cardiomyopathies were defined as "diseases of the myocardium associated with cardiac dysfunction", and they were classified according to anatomy and physiology into the following four types: Hypertrophic cardiomyopathy, HCM; Dilated cardiomyopathy, DCM; Restrictive cardiomyopathy, RCM; Arrhythmogenic right ventricular cardiomyopathy, ARVC, and Unclassified cardiomyopathies. Cardiomyopathies that are associated with specific cardiac or systemic disorders generally fall into one of these categories. These include ischemic, valvular, hypertensive, inflammatory, toxic, mitochondrial, neuromuscular, metabolic, and inherited disorders (FIGURE 1) [2].
These disorders have been also indicated as "phenocopies" The term "specific cardiomyopathy" or "phenocopies" was probably the first important step toward a new classification, reflecting the fact that the genetic basis of the cardiomyopathies was being elucidated. Indeed, over the last two decades, clinical and molecular insights helped to better understand aetiology and management of cardiomyopathies. Many disorders considered before as "idiopathic" or "primary" disorders have been associated to specific genetic or non genetic defects, clinical features and outcome.
The American Heart Association Classification
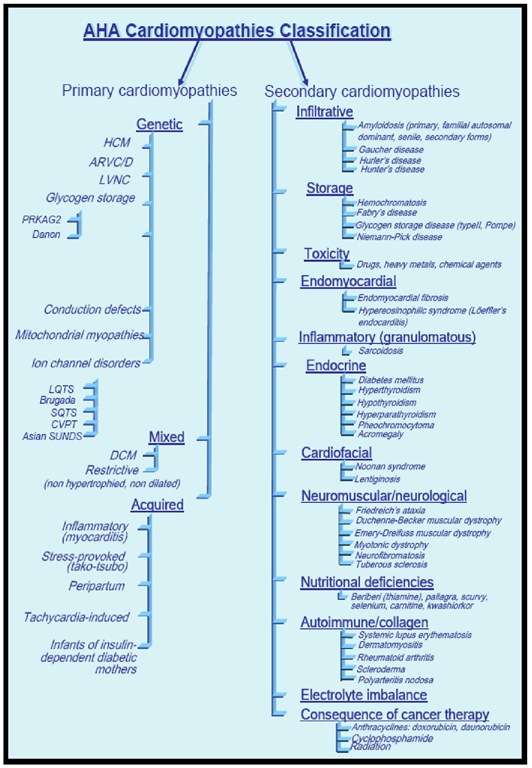
Figure 1. The American Heart Association classification of cardiomyopathies.
In 2006, an expert committee of the American Heart Association proposed the following definition of cardiomyopathies: "Cardiomyopathies are a heterogeneous group of diseases of the myocardium associated with mechanical and/or electrical dysfunction that usually (but not invariably) exhibit inappropriate ventricular hypertrophy or dilatation and are due to a variety of causes that frequently are genetic. Cardiomyopathies either are confined to the heart or are a part of generalized systemic disorders, often leading to cardiovascular death or progressive heart failure-related disability."
They also proposed a new scheme of classification, in which the term "primary" is used to describe diseases in which the heart is the sole or predominantly involved organ and "secondary" to describe diseases in which myocardial dysfunction is part of a systemic disorder. Primary cardiomyopathies have been sub-classified in genetic forms, mixed forms (genetic and non genetic), or acquired forms.
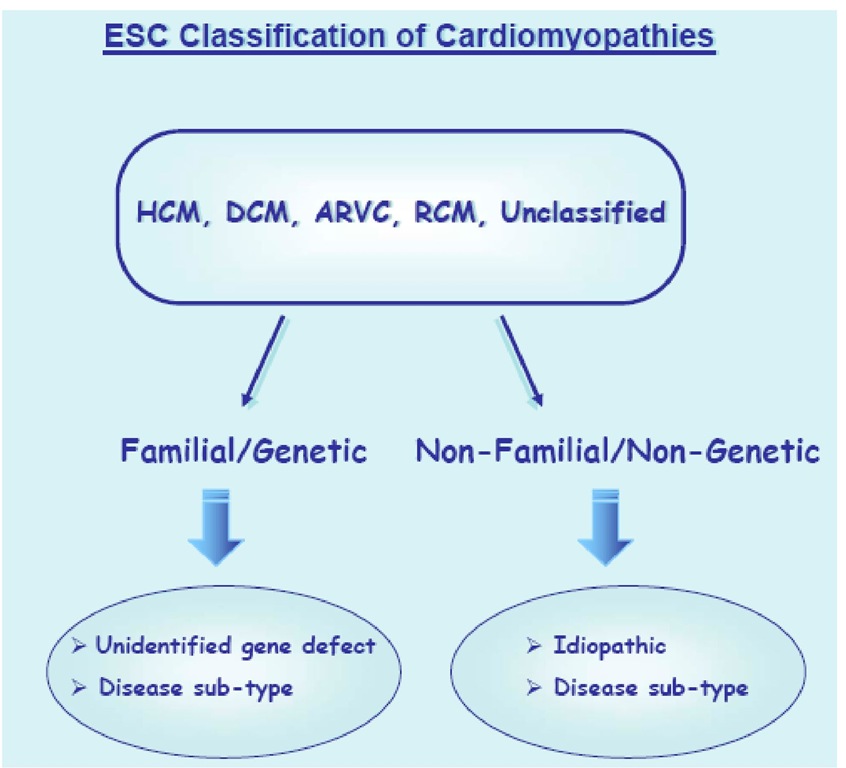
Figure 2. The European Society of Cardiology classification of cardiomyopathies.
The main departure of the proposed AHA Scientific Statement definition from previous classifications is the inclusion of the ion channelopathies as primary cardiomyopathies, despite the absence of gross structural abnormalities (FIGURE 2) [3].
The European Classification
Figure 3. The World Health Organization classification of cardiomyopathies.
In 2007, the Working Group on Myocardial and Pericardial Diseases of the European Society of Cardiology proposed an update of the WHO/ISFC classification, defining cardiomyopathy as: "A myocardial disorder in which the heart muscle is structurally and functionally abnormal in the absence of coronary artery disease, hypertension, valvular disease and congenital heart disease sufficient to explain the observed myocardial abnormality." Cardiomyopathies are grouped into specific morphological and functional phenotypes: each phenotype is then subclassified into familial/genetic and non-familial/non genetics forms. Like the 2006 AHA proposal, it focuses on the established morphological distinctions (hypertrophic cardiomyopathy, dilated cardiomyopathy, arrhythmogenic right ventricular cardiomyopathy, restrictive cardiomyopathy). Unlike the AHA classification, heart disease secondary to coronary heart diseases, valvular and congenital disorders are not included. Channelopathies are excluded as well (FIGURE 3) [4].
Genetics of Cardiomyopathy: A Long Way to Go
From "One Gene-One Disease"…
In 1989, Christine and Jon Seidman and co-workers reported the first association between an inherited gene defect and a primary cardiomyopathy [5]. In a subsequent study, they reported the first beta myosin missense mutation in a French Canadian family with hypertrophic cardiomyopathy, leading to the equation "one gene=one disease" [6].
.To "One Disease-Many Genes".
Since then, substantial progress has been made in elucidating further (sarcomeric and non sarcomeric) gene defects in cardiomyopathies. To date, over 450 mutations in sarcomere protein genes (thick and thin filaments) have been identified in patients with HCM: P-myosin heavy chain (chromosome 14); cardiac troponin T (chromosome 1); cardiac troponin I (chromosome 19); troponin C (chromosome 3); a-tropomyosin (chromosome 15); cardiac myosin-binding protein C (chromosome 11); the essential and regulatory myosin light chains (chromosomes 3 and 12, respectively); cardiac actin (chromosome 15), titin (chromosome 2), and a-cardiac myosin heavy chain (chromosome 14). Some patients with sporadic disease, have similar genetic abnormalities as those with familial disease. De novo mutations in cardiac myosin binding protein-C, cardiac beta-myosin heavy chain, cardiac troponin T or alpha-tropomyosin genes have been found in isolated case reports of individuals with sporadic HCM [7].
HCM was then characterized as a disease of the sarcomere. As a consequence, DCM was indicated as a disease of the cytoskeleton and extracellular matrix, ARVC of the desmosome, and so on.
.To "One Disease-Many (Different)Genes"…
However, this "systematic" view became, again, in contrast with the rapidly growing genetic knowledge. Indeed, mutations in sarcomeric genes account for approximately 50-60% of all cases of HCM, which is the most know form of cardiomyopathy both in the clinical and genetic setting, so far. The absence of sarcomeric gene mutations in the remaining HCM population seems to be related to a shortcomings in current mutation detection methods and strategies, or to be a result of disease-causing mutations in yet unidentified genes [7].
Rare causes of HCM have been associated with mutations in sarcomere-related protein genes (myosin light chain kinase, muscle LIM protein, LIM binding domain 3, telethonin, vinculin and metavinculin, caveolin, titin, a-actinin 2, myozenin 2, and junctophilin 2), or functional genes (phospholamban, RAF-1) [7].
Another possible explanation for the low proportion of cases thought to be caused by sarcomere gene mutations is that the population of hypertrophic cardiomyopathy without sarcomeric gene mutations may carry one of the several diseases that mimic the phenotypic expression of sarcomeric hypertrophic cardiomyopathy (the so called "phenocopies" of the disease), including metabolic-mitochondrial-neuromuscular diseases, inherited syndromes, and chromosomal abnormalities [7].
Mutations in the genes encoding the gamma-2 regulatory subunit of adenosine monophosphate (AMP)-activated protein kinase (PRKAG2) and lysosome-associated membrane protein 2 (LAMP2) have been associated with hypertrophic cardiomyopathy in association with Wolff-Parkinson-White (WPW) syndrome [7,8]. Similar to PRKAG2 and LAMP2, Fabry’s disease, an X-linked lisosomial disorder, can express predominant cardiac features of left ventricular hypertrophy. Over the years, mutations in GLA-encoded alpha-galactosidase A have been found in patients with this multisystem disorder [7,9]. Friedreich ataxia, an autosomal-recessive disease involving sclerosis of the spinal cord, is often associated with cardiomyopathies, and its cardiovascular manifestations may precede the neurological symptoms by up to a decade in some cases [7,10]. Chromosome Abnormalities, including Down syndrome and trisomy 18, Autosomal Dominant Cardiofacial Disorders (Noonan syndrome, LEOPARD syndrome, Cardiofaciocutaneous syndrome Costello syndrome) or Phakomatoses (Neurofibromatosis, Tuberous sclerosis) have been associated with a variety of cardiac defects, including hypertrophic cardiomyopathy [7,11-14].
…To "One Gene-Many Diseases"
The major cardiomyopathies are genetically heterogeneous diseases for which the causative genes are partially overlapping. The evidence that a complex of genes (i.e. sarcomeric protein genes) may be responsible of a "spectrum" of different phenotypes, including HCM, DCM, RCM and recently LVNC, represent a further step in the knowledge and understanding of cardiomyopathies[15]. The identification of sarcomeric mutations in familial LVNC, and an alpha-actin mutation in HCM with LVNC and atrial septal defect together with the observation of late onset LVNC in a Duchenne patient, suggests that the aetiology of LVNC extends beyond an arrest in embryonic cardiac development (i.e. the possibility of late onset LVNC[16,17]. The current findings expand the genetic heterogeneity of LVNC, and the identification of sarcomeric defects in familial LVNC suggests that LVNC may be part of a cardiomyopathy spectrum including HCM, RCM, and DCM[16,17].
Whether this means that these are different diseases or rather different manifestations (phenotypes) of the same pathological mechanism is presently not clear.
… Or to "One Gene-Many (Different)Diseases"…
It is now evident that a large number of mutations in different genes, albeit largely within the same class, could cause the same phenotype. Moreover, mutations in one gene could cause multiple phenotypes, as best illustrated in the case of lamin A/C, whereby mutations can cause 13 different diseases, including DCM, conduction defects, Emery Dreifuss muscular dystrophy, familial partial lipodystrophy, premature aging, axonal neuropathy, and insulin resistance [18]. In addition, SCN5A (sodium channel) gene mutations may cause phenotypes that combine features of LQT3 (Long QT Syndrome 3), Brugada syndrome, conduction disease and dilated cardiomyopathy ("one gene-different diseases") [19].