1. Introduction
Hidden within an individual’s genomic sequence are the genetic instructions for the entire repertoire of cellular components that determine the complexities of biological systems. Unraveling genomic structure and characterizing the functional elements from within the code will allow connections to be made between the genetic blueprint, transcribed information, and the resulting systems biology, and will, in turn, accelerate the exploration of the biological sciences.
As pointed out in a recent review (Shendure etal., 2004), the vast majority of known DNA sequence data to date have been generated using the Sanger-based sequencing method. However, genotyping (see Article 77, Genotyping technology: the present and the future, Volume 4) has been the tool most widely chosen for genetic exploration because the cost of sequencing individual genomes remains prohibitively expensive (recent estimates place the cost of sequencing a human genome in the region of tens of millions of US dollars). Technological advances in DNA resequencing, leveraging the availability of the consensus genome sequence for almost 200 species (http://www.intlgenome.org/viewDatabase.cfm), are transforming throughput and costs. An improvement in the region of four to five orders of magnitude over current sequencing costs is no longer an unrealistic prospect.
High-throughput sequence analysis, using capillary-electrophoretic separation and four-color fluorescent detection in instrument systems (such as the Applied Biosystems 3700/3730 and Amersham Biosciences MegaBACE 1000/4000), has been deployed successfully in factory-scale operations, largely within public-funded organizations, to sequence the human genome and that of many other species (see Article 5, Robotics and automation, Volume 3). Improvements in these systems continue to deliver incremental (maybe as high as 10-fold) increases in throughput and cost reductions. But these do not address the fundamental need for a transformation in cost-effectiveness that would be necessary for sequencing on a genome-wide scale to become a routine undertaking. Reagents are currently a highly significant cost element in current sequencing approaches and therefore a key target in cost-reduction approaches.
An initiative to address this key cost factor is being taken by the laboratory of Richard Mathies at U.C. Berkeley (Paegel et al., 2003). Mathies’ lab is working to achieve this goal by seeking to create an integrated microfabricated device that couples clone isolation, template amplification, Sanger extension, purification, and separation in a single microfluidic device. They highlight the development (in their lab and that of other workers) of highly parallel microfabricated capillary-array electrophoresis analyzers, nanoliter-scale DNA purification and amplification reactors, and microfluidic cell sorters and cytometers to support the feasibility of creating such an integrated microfabricated device. They calculate that the processing time could be reduced by 10-fold and reagent consumption by 100fold, compared to the current state of the art. To go beyond this in cost reduction requires a fundamentally different strategy.
2. New sequencing approaches
There are several emerging sequencing technologies that aspire to ultralow cost, ultrahigh-throughput capabilities. Shendure et al. (2004) have classified these methods broadly into five different groups: microelectrophoretic methods (such as the work of Mathies and colleagues referred to above), hybridization, cyclic-array sequencing on amplified molecules, cyclic-array sequencing on single molecules, and real-time methods. While each of these approaches has potential to make the necessary breakthrough in technology, it is too early to predict whether expectations will be fulfilled. Yet, for some, and in particular the single molecule array-based approaches, recent developments have continued to stimulate community interest in sequencing technologies that have the capability to analyze entire genomes very quickly at an affordable price. This is particularly so for the human genome and the aspiration to achieve the so-called $1000 human genome concept (Zimmerman, 2004).
3. Single molecule-based approaches
Analysis at the single molecule level is challenging, yet it offers substantial advantages not only over conventional sequencing but also over other emerging technologies. Recent progress in the development of highly efficient strategies that dramatically reduce reagent consumption during analysis is bringing the routine analysis of whole-genome variation at the sequence level closer to reality. Methods under development, as reviewed by Smith (2004), fall into three main categories:
• Single molecule separation: Elongation of large fragments of genomic DNA that has been tagged with fluorescently labeled probes bound at specific sites, such as that being developed by OpGen (http://www.opgen.com) and US Genomics (http://www.usgenomics.com). The molecules can be analyzed at high speed, and this taken with the currently low (>1200 bp) resolution makes such techniques suited to mapping rather than sequencing.
• Arrays of cloned single molecules: Sequencing techniques related to high-density arrays of “colonies” of identical copies of template amplified from a single molecule, either immobilized on a solid surface (e.g., Solexa’s cluster array developed by former Swiss company Manteia) dispensed into a very high density microtiter plate (e.g., 454 Life Sciences http://www.454.com), or amplified in a thin polyacrylamide gel matrix on a slide (e.g., Church’s group at Harvard; Mitra and Church, 1999). • Single molecule arrays: Single molecule analysis in an array-based format to generate a massively parallel approach to sequencing, as pioneered by Solexa (http://www.solexa.com).
4. Single molecule array-based approaches
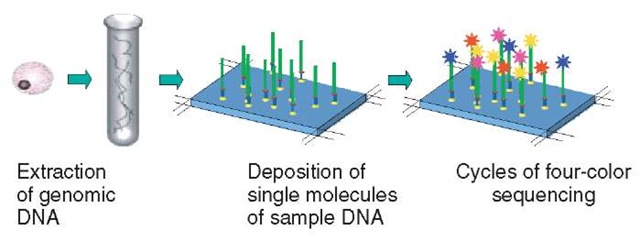
Single molecule array-based approaches are characterized by a number of distinct advantages over other technologies (Figure 1). In addition to minimal sample preparation and a novel sequencing chemistry, the rapid detection of single, fluorescently labeled dye molecules with very high signal-to-noise ratio is a critical feature of Solexa’s Single Molecule Array™ (SMA) technology. The technology is massively parallel with an estimated 100 million single molecules of DNA sample template, dispersed randomly, per square centimeter of array. In the presence of a proprietary polymerase, specially designed nucleotides act as reversible terminators of sequencing so that, at each cycle, only a single base of DNA template is sequenced. Each of the four nucleotides is labeled with a distinguishable fluor and detected using a four-color detection system. Once the base has been identified, the block to further extension is relieved and the fluorescence removed so that the next cycle can be performed. Development of reversibly terminating nucleotides, by limiting each cycle to a single incorporation, overcomes the problem encountered by other approaches of having to decipher homopolymeric sequences and increases the accuracy of incorporation by the polymerase as all four nucleotides are present in the sequencing reaction.
The number of cycles of sequencing is dictated by the size of the genomic template that is under investigation. For example, with human resequencing, each template is sequenced to a length of 25 to 30 bases, derived from an analysis by Solexa of the human genome that revealed that unique alignment requires a read length of approximately 20 bases. Software aligns the n-mer reads against the reference sequence of the genome to identify a large part of the variation between the individual’s DNA and the reference sequence. In this way, unknown SNPs as well as known SNPs can be detected and typed simultaneously at the same time as gathering data to determine haplotype structure and patterns of linkage disequilibrium. The approach is universally applicable to any organism for which a reference sequence is available, and shorter read lengths can be used where the genome, or a genome entity, such as a single chromosome, is less complex.
Figure 1 Single molecule array sequencing. Arrays of single molecules are created by binding randomly fragmented genomic DNA to a chip surface as primed templates. Addition of fluorescently labeled nucleotides and DNA polymerase allows sequence determination.
As the cost of sequencing per se is reduced, sample preparation will account for a significant proportion of total costs (see Article 4, Sequencing templates -shotgun clone isolation versus amplification approaches, Volume 3). Performed in a single reaction, the SMA approach does not require costly or time-consuming preparation, such as PCR amplification or cloning target DNA into bacteria. Another important advantage of SMA is the requirement only for very small quantities (picograms) of DNA starting material. This not only avoids averaging effects of using large samples, masking what is really happening in a biological system but also avoids representational bias by minimizing sample processing. These features, together with a dramatic reduction in reaction volume, combine to revolutionize the economic landscape of sequencing. Once viable economically at large scale, whole genome resequencing of each sample will enable true whole-genome association studies.
A critical consideration is that these new approaches will produce an unprecedented quantity of data, which will have to be processed, annotated, and applied. This will require an entirely new set of skills, systems, and databases, which, it is anticipated, will create an entirely new field of genomics. To this end, Solexa is working with the groups of Ewan Birney and Richard Durbin at the European Bioinformatics Institute and the Wellcome Trust Sanger Institute, respectively, to extend and advance the Ensembl system (http://www.ensembl.org) to manage, query, and visualize multiple whole-genome sequence data sets. Furthermore, as a second strand to this project, statistical methods and tools are being developed with David Balding and colleagues at Imperial College London to allow epidemiological studies to exploit whole-genome data to localize gene effects involved in disease susceptibility and drug metabolism.
5. Arrays of cloned single molecules
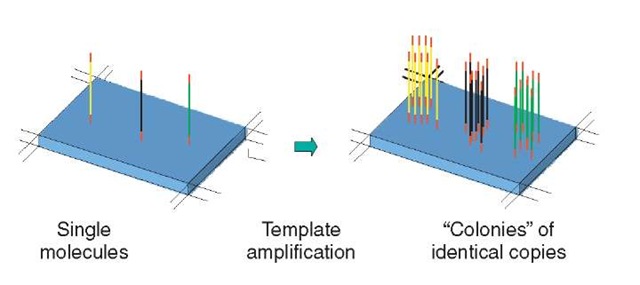
There is a group of related techniques that seeks to overcome the high sensitivity required to analyze individual single molecules, by creating a high-density array of “colonies” of identical copies amplified from a single molecule (Figure 2). Church’s group at Harvard (Mitra and Church, 1999; Mitra et al., 2003) carry out PCR amplification in a thin polyacrylamide gel matrix on a slide to constrain lateral diffusion, thereby creating colonies of PCR products; they coined the term “polonies” to describe these. A related strategy, Manteia approach (Adessi et al., 2000), involved amplification of single-molecule templates immobilized on a solid surface. 454 Life Sciences (Leamon et al., 2003) have dispensed single molecules into a very high density microtiter plate, such that each 75-picoliter well contains no more than one molecule, and then carried out amplification. The sequences of several viral and bacterial genomes have been determined using this approach.
Figure 2 Arrays of cloned single molecules. Single molecules are amplified in a spatially defined way such that a large number of identical copies of each are generated in isolated “colonies”. These colonies can then be subjected to sequencing in situ.
These arrays of cloned single molecules are then subjected to sequencing using, for example, DNA polymerase-based incorporation of labeled nucleotides and fluorescence detection or pyrosequencing (http://www.pyrosequencing.com). The use of cloned single molecules facilitates detection by yielding a higher signal than an individual single molecule. In principle, this allows detection instrumentation that is relatively less sophisticated and less costly to be employed. A somewhat greater level of inefficiency in the sequencing biochemistry or loss of templates through the process can be tolerated, as the signal is derived from a large number of molecules. Balanced against these considerations, cloned single molecules can introduce problems owing to the individual molecules in a colony becoming out of phase with one another during the sequencing process and therefore creating high backgrounds and spurious signals. Other issues are the complexity and effort involved in generating the cloned array and the potential for the sequence representation of the sample not to be faithfully preserved.
6. Applications
By applying different simple methods of sample preparation and downstream analysis algorithms to the core technology, the range of capabilities of SMA technology is extended. SMA can be applied either to resequence whole genomes or to the same reproducible, specific genome sequence from several different individuals to provide a particular set number of SNP loci (e.g., a particular subset of, say, 1 million SNPs) for mapping traits (Bennett, 2004; see also Article 11, Mapping complex disease phenotypes, Volume 3 and Article 17, Linkage disequilibrium and whole-genome association studies, Volume 3).
The primary application focus is on basic research, both in academia and in industry, where this breakthrough technology is anticipated to stimulate a new wave of research activity enabled by the newfound ability to measure variation comprehensively across whole genomes (see Article 68, Normal DNA sequence variations in humans, Volume 4). The technology will stimulate new methods of applying knowledge of individual variation with wide-ranging applications, such as is in functional/comparative genomics (see Article 48, Comparative sequencing of vertebrate genomes, Volume 3), exploration of microbial diversity for the agricultural biology field, pathogen identification (see Article 49, Bacterial pathogens of man, Volume 4), transcriptome characterization and in particular of alternative splice variants, genotype-phenotype correlations, human and animal disease association (see Article 57, Genetics of complex diseases: lessons from type 2 diabetes, Volume 2), pharmacogenomics, the development of new molecular diagnostics and drugs, and in personalized medicine. This process will begin largely in the major government-funded and not-for-profit-funded research institutes, leveraging the strong political will that exists to see real human health benefits from the large investment already made in genetics, and in particular in the Human Genome Project (see Article 24, The Human Genome Project, Volume 3) and its various ramifications.
7. Concluding remarks
Single molecule array-based sequencing technology has the potential to transform the economics of DNA sequencing by allowing the sequence of hundreds of millions of individual molecules to be determined rapidly in parallel. The approach drastically reduces, and at best obviates, the need for sorting, cloning, and amplification of genomic DNA samples with the consequential reduction in laboratory preparation and reagent overheads. Together, these facets of single molecule array-based sequencing will allow sequencing of large entities genomic, including whole genomes, at costs several orders of magnitude below current levels. For human genetics, next-generation technologies such as SMA offer the potential to achieve the much sought after $1000 human genome goal.