1. Introduction
High-quality DNA is essential to obtaining the greatest success in DNA sequencing. Sequencing quality DNA should contain the lowest possible level of contaminating host DNA, have consistent yields from sample to sample, and be in sufficient quantities to perform multiple sequence reactions. Many thousands of clones are required whether a whole-genome shotgun or clone-by-clone approach is taken so the DNA isolation method must be amenable to automation for a high-throughput sequencing process.
To produce sequencing templates, random fragments of DNA are inserted into cloning vectors and propagated in Escherichia coli. Individual clones are cultured in 0.5-2 ml volumes and the episome containing the clone DNA is extracted and purified. Each method differs in the quality of DNA produced, the length of time and cost to perform, and its suitability to automation. Variable DNA concentrations from sample to sample make it difficult to optimize sequencing reactions. DNA isolation may be one of the most basic and routine molecular biology techniques but it is one of the most important for success.
Traditional extraction methods involve chemical isolation of the subclone DNA, taking advantage of the mass difference between the plasmid and chromosomal DNA. Some techniques further involve immobilization of the plasmid to a solid structure. DNA amplification methods circumvent the cell growth step, allowing DNA isolation to proceed directly from bacterial colonies or glycerol stocks. Table 1 shows a comparison of three common methods of DNA preparation for plasmid clones – alkaline lysis (Birnboim and Doly, 1979), solid-phase reversible immobilization (SPRI) (Hawkins etal., 1994), and rolling circle amplification (RCA) (Dean et al., 2001).
2. Cloning and sequencing vectors
Bacterial artificial chromosomes (BACs) are probably the most common clone type used today for genomic library construction. Libraries in BAC vectors such as pBACe3.6 were used for many of the model organism sequencing projects including human and mouse (Osoegawa et al., 2000). The BAC cloning vectors are based on the naturally occurring F-factor found in E. coli and they are maintained as supercoiled circular episomes within the bacteria, usually with a single copy per cell. The vector to insert ratio for BACs is very good. Inserts up to 300 kilobases (kb) can be stably introduced into the approximately 8-kb vector. Fosmid vectors (Kim et al., 1992) also contain the E. coli F-factor, but only 40 kb can be stably maintained in these vectors.
Table 1 Comparison of some popular DNA preparation methods
| DNA isolation method | |||
| Alkaline lysis | SPRI | RCA | |
| Time to prepare 96
templates Yield |
18 h, including overnight
culture. 7 |g from 1.5-ml culture |
16.5 h, including overnight
culture. 4 |g from 1.5-ml culture |
4.5h
1.5 |g from single colony |
| Quality of DNA Sample-to-sample variability | Good High | High Low | High
No variability |
| Number of liquid-handling
steps |
8 | 7 | 2 |
| Plastic ware | Culture plate Microwell plate | Culture plate Microwell plate | Microwell plate |
| Equipment required | Incubator
Centrifuge Vortex |
Incubator Magnet | Water bath |
| Ease of automation
Key reagents |
Not easily automated
Bacterial growth media GTE NaOH SDS KOAc Ethanol |
Easily automated Bacterial growth media SprintPrep buffer Isopropanol Ethanol | Easily automated Denature buffer TempliPhi Premix |
The most common sequencing vectors by far are the double-stranded plasmids, usually the high copy number pUC-based plasmids (Yanisch-Perron et al., 1985). With double-stranded vectors, sequence data from both forward and reverse strands can aid assembly of the genome or clone in question (Roach et al., 1995). The previously favored single-stranded bacteriophage M13-based vectors are now usually used only for regions that are not stably maintained in pUC plasmids (Chissoe et al., 1997). Increased sequence readlength from improvements in sequencing chemistry and instrumentation has allowed an increase in the typical subclone insert size. Sequence readlengths of 700-800 base pairs (bp) are not uncommon and so an average insert size of 2-4 kb is now routinely used for shotgun libraries in plasmid vectors.
3. Traditional plasmid DNA isolation techniques
When Wilson et al. (1992) described the methods involved in sequencing a 95-kb section of the mouse genome, the processing of 24M13-based subclones took one individual almost a day. With current levels of automation, thousands of subclones can be prepared per day, with human involvement reduced to loading and unloading of microwell plates and reagents.
The most common method for extraction of plasmid DNA from E. coli cells is still alkaline lysis. This method takes advantage of the mass differences between plasmid and chromosomal DNA. Bacteria are lysed by treatment with a solution containing sodium dodecyl sulfate (SDS) (CAS # 151-21-3) that denatures the proteins, and sodium hydroxide (NaOH) (CAS # 1310-73-2) that denatures chromosomal DNA. The mixture is neutralized with potassium acetate (KOAc) (CAS # 127-08-2) and the supercoiled, plasmid DNA reanneals rapidly due to its secondary structure and smaller size. The chromosomal DNA and proteins form a solid precipitate with the insoluble potassium salt and SDS and pellet under centrifugation. The plasmid is further purified from the supernatant by alcohol precipitation and washing.
An alternative method to alkaline lysis is the boiling miniprep (Holmes and Quigley, 1981). The cells are lysed by treatment with lysozyme (CAS # 1265088-3) and heating in the presence of Triton X-100 (CAS # 9002-93-1) and sucrose (CAS # 57-50-1). This procedure releases the plasmid DNA but not the chromosomal DNA from the cell. Centrifugation pellets the cell debris including most of the chromosomal DNA, leaving the plasmid DNA in the supernatant, which is further purified by alcohol precipitation. This method is quicker than alkaline lysis, but the quality of the DNA is lower, having higher chromosomal DNA contamination and more variable yield.
Variability in yield can have a dramatic effect on sequence DNA quality. It is difficult to optimize sequencing reactions when the DNA templates vary widely in concentration. In addition, capillaries in DNA analysis systems can be adversely affected by excessive amounts of DNA in the samples. Sequencing capillaries vary in the range of DNA that they can tolerate, and the type of sequencing instrument should be a consideration when deciding which isolation method to use.
One of the main advantages of both the alkaline lysis and boiling methods is cost. The reagents are inexpensive and easily obtainable and no special equipment is needed, beyond a centrifuge. Once the overnight cell growth is complete, the procedures are fairly quick; two 96-well plates of cultures can be processed in a few hours by a single technician. DNA quality is usable, but probably the lowest of the methods that will be discussed – a chromosomal DNA contamination level of 5-10 % can be expected. As these methods involve centrifugation, they are difficult to automate which is vital for either a cost effective or high-throughput operation.
4. Filter-based purification methods
Most of the commercially available plasmid purification products, such as R.E.A.L™ (rapid extraction alkaline lysis) Prep 96 Plasmid Kit (Qiagen Inc.), begin with the alkaline lysis procedure but differ in the purification step. Following cell resuspension, lysis, and neutralization, the lysate is passed through a membrane that binds the plasmid DNA. The plasmid DNA is washed and then eluted with water or Tris-EDTA (TE) buffer (CAS # 77-86-1 and 139-33-3). These so-called bind-wash-elute products usually use glass fiber membranes or glass beads that bind DNA in the presence of a chaotropic salt such as guanidine hydrochloride (CAS # 50-01-1). The lysate is usually drawn through the membrane using a vacuum manifold.
These methods eliminate some of the centrifugation steps in the alkaline lysis protocol, making them more amenable to automation and are available in single, 96-well and 384-well formats. Without the alcohol precipitation step, the methods are generally quicker than standard alkaline lysis methods. A 96-well plate of minipreps can be prepared from grown cultures in 45 min. DNA purity with these products is usually higher than with the standard alkaline lysis procedure but the overall cost is also increased owing to the additional filter plates required.
5. Alternative plasmid purification methods
5.1. Solid phase reverse immobilization (SPRI)
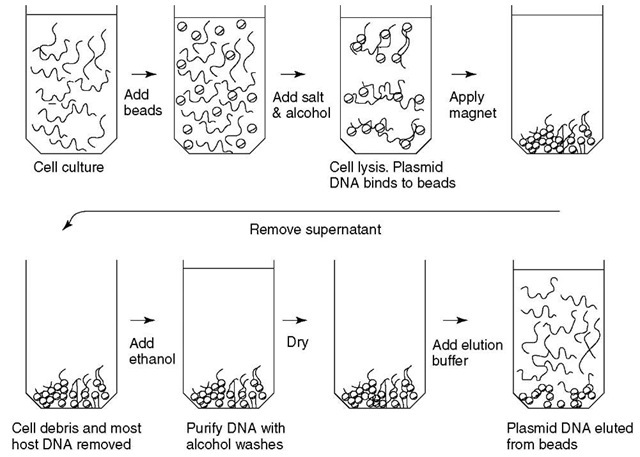
Technologies that use physical isolation of DNA instead of chemical isolation are commercially available. One such method, SPRI, is used in the SprintPrep™ and CosMCPrep™ DNA purification kits (Agencourt Biosciences Corp.). Carboxyl-coated magnetic beads in the presence of high polyethylene glycol (PEG), alcohol, and salts bind plasmid DNA from lysed bacterial cultures (Figure 1). Cell pelleting and resuspension steps are eliminated by using magnetic separation. Beads with absorbed DNA are washed with ethanol (CAS # 64-17-5) to remove contaminants, then the plasmid DNA is eluted from the beads with water. As this method requires neither centrifuge nor vacuum manifold, it can easily be automated. This method is the quickest of the ones discussed here. A 96-well plate of bacterial cultures can be processed in about 20 min.
5.2. Rolling circle amplification
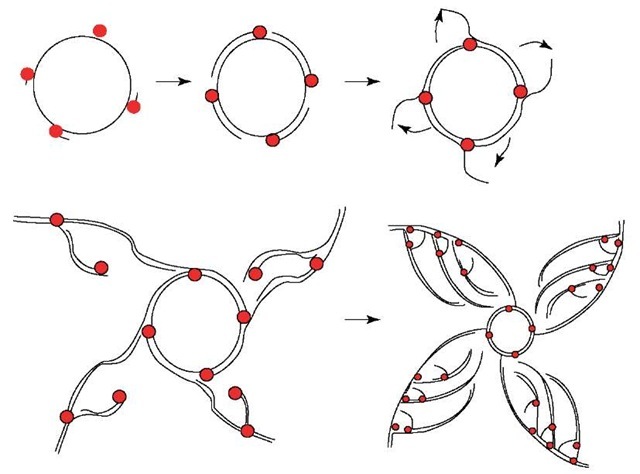
All of the methods discussed so far employ overnight cell growth to propagate plasmid-containing cells and thus amplify the cloned DNA. These methods are effective when high copy-number vectors are used. An alternative strategy is to use multiply primed RCA. This method uses a highly processive, strand-displacing DNA polymerase to amplify the plasmid DNA directly from bacterial colonies, eliminating the need for overnight culture. TempliPhi™ DNA Sequencing Template Amplification kits (GE Healthcare) exploit this technology. Over 10 000-fold amplification can be achieved in as little as 4 h using random hexamer primers that initiate multiple replication forks (Figure 2).
The key to the technology is the DNA polymerase from bacteriophage Phi29. This DNA polymerase is highly processive, incorporating more than 70 000 nucleotides in a single binding event (Blanco et al., 1989). RCA is an isothermal reaction and does not require cycling to denature the DNA strands for the next round of amplification as in polymerase chain reaction (PCR). When the enzyme encounters a nontemplate strand, it simply displaces it, generating single-stranded DNA available for further primer annealing. This leads to exponential amplification of both strands. Phi29 DNA polymerase has a 3′-5′ exonuclease activity, giving it an error rate of only 1 in 106-107 (Esteban et al., 1993), approximately 100 times lower than Taq DNA polymerase (Dunning et al., 1988). The product of the RCA process is double-stranded concatamers of the input DNA sequence (Figure 3). Approximately 80% of the product can be digested with restriction endonucleases generating unit-length DNA fragments (Dean et al., 2001).
Figure 1 Schematic of SPRI plasmid isolation procedure. Paramagnetic beads are added to bacterial culture. Cells are lysed, and plasmid DNA binds to paramagnetic beads in the presence of isopropanol and salts. Immobilized plasmid DNA is further purified by ethanol washes. DNA is eluted from the beads with water
There are a number of advantages to this technique. Speed is an obvious one; sequence ready DNA can be prepared in under 5 h, directly from colonies, with only 20min of hands-on time for a 384-well plate of templates. Another is consistency of yield. Properly formulated, RCA is an exponential reaction, terminating only when all the nucleotides in the reaction mixture have been exhausted. Every reaction yields the same mass of DNA product, making optimization of downstream sequencing processes much simpler and more reliable than with other methods. In addition, the amplification product can be used directly in sequencing reactions without any further purification. It is not necessary to remove the excess hexamers prior to sequencing as they will not participate in the sequencing reaction owing to their lower melting temperature compared to sequencing primers. The one major disadvantage may be cost, the reagents being more expensive than those used in the alkaline lysis procedure but on par with other commercial plasmid purification methods. The increase in reagent cost may be offset by savings in the time, labor, and space, which can be achieved by the elimination of the bacterial growth and many liquid handling steps.
Figure 2 Schematic of rolling circle amplification. Random hexamers bind to the circular template, generating multiple replication forks. Phi29 DNA polymerase displaces the nontemplate strand, making them available for further primer binding. The amplification product is double-stranded tandem copies of the starting circle
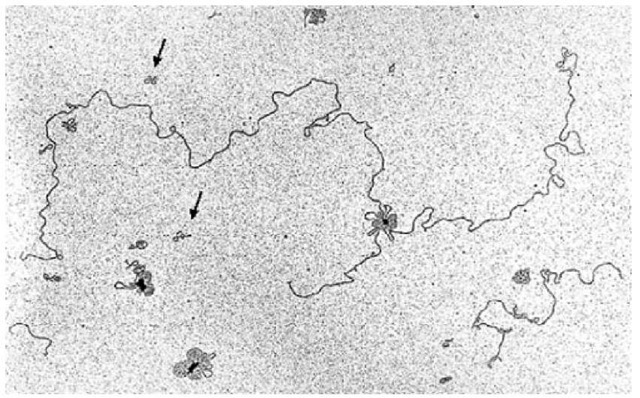
Figure 3 Electromicrograph of plasmid DNA amplified by rolling circle amplification. Image shows RCA products after 5 min amplification. Arrows indicate unit-length (nonamplified) plasmid molecules
5.3. Colony PCR
Another method that bypasses culture growth is colony PCR (Gussow etal., 1989). A colony is simply picked into a PCR cocktail containing primers in the flanking vector sequence designed to specifically amplify the entire insert. It is often necessary to purify the PCR product from the primers and excess nucleotides to prevent them from interfering in the sequencing reaction. Kits such as ExoSAP-IT™ (usb Corp.) contain E. coli exonuclease I and shrimp alkaline phosphatase to remove the single-stranded primers and free nucleotides. Colony PCR is a quick and simple method but has not been extensively used because of the amplification errors that can be introduced by the PCR process. The guidelines set out by the major sequencing centers on finishing DNA sequence (G16 Finishing Standards for the Human Genome Project – Version September 7, 2001 http://www.genome.wustl.edu/Overview/finrulesname.php?G16=1) limit the amount of the genome that can have sequence coverage only from PCR products, and any sequence derived from PCR products must be annotated. Despite the error rate, this method can be useful for quick colony screening.
6. Factors affecting plasmid yield
Plasmid yield is dependent on many factors including type of plasmid, (high or low copy number), size of plasmid, and E. coli host strain. For instance, copy number can vary from approximately 1000 for pUC vectors down to less than 10 for vectors with functional copy-control. Plasmid size should be taken into account when choosing an isolation procedure. Methods such as the boiling miniprep that rely on plasmid DNA being released from the cell when lysed are not suitable for large insert plasmids (>10kb) as the plasmids get withheld along with the chromosomal DNA. This should also be a consideration for RCA where the harsher lysis conditions required to release the plasmid may release host DNA, which will be amplified in addition to the plasmid DNA. This is especially true for large vectors such as fosmids (see below). Optimization of PCR conditions may be required for colony PCR of plasmids with inserts larger than about 2 kb. Different PCR conditions may be required for vectors with different sized insert.
For alkaline lysis-based methods, including the filter-based methods, ideal yield from a 1.3-ml culture of a high-copy plasmid such as pUC is approximately 7 |g, although there is considerable sample to sample variability. With RCA, the yield is dependent on the amount of nucleotide in the reaction. Currently, two DNA amplification kits are available commercially from GE Healthcare that produce either 1.5|g in about 4h or 3.5|g in 18h. These reactions can be scaled up or down if more or less DNA is needed. With the SprintPrep DNA purification kit from Agencourt Biosciences, 150 |l of culture yields about 400 ng of plasmid DNA.
7. Preparation of M13-based vector DNA
Purification of M13-vectors is much simpler than for plasmids because the M13 phage particles are released into the growth media. Cells are pelleted by centrifu-gation, and the phage particles precipitated from the supernatant using PEG (CAS # 25322-68-3) and salt. The M13 DNA is released from the coat protein during the denaturation steps of cycle sequencing so no further purification is necessary for sequencing quality M13 DNA. If ultra pure M13 DNA is required, the DNA can be further purified by alcohol precipitation and washing. RCA and PCR can be used to amplify M13 templates directly from plaques. The product of both methods is double stranded and can immediately be sequenced from both the forward and reverse strands.
8. Preparation of fosmid and BAC DNA
The difficulty with purification of large vector constructs is twofold. First, they are usually present in only one or two copies per cell (although high copy-number vectors are recently available) and second, they are much larger than subclones, making purification based on size more difficult. There are many different protocols available for the isolation of BAC DNA, depending on the purity of DNA required. For sequencing purposes, some chromosomal DNA contamination is acceptable, but if the same DNA is to be used for fingerprinting, then a method that gives higher purity DNA may be required.
Alkaline lysis is used to purify BACs and fosmids as they are maintained in E. coli as supercoiled episomes. Owing to the increased size of BAC and fosmid constructs compared to plasmid subclones, some BAC DNA inevitably complexes with the SDS, protein, and chromosomal DNA, resulting in low yields. Some protocols allow the samples to stand for 30min after the addition of the potassium acetate, presumably to allow time for the large construct DNA to reanneal. Depending on the level of purity required, either alcohol precipitation or cesium chloride gradient centrifugation can be performed following neutralization to improve DNA quality.
Filter-based methods are also available for purification of large constructs. As for plasmid purification kits, they are based on the alkaline lysis method followed by membrane purification in place of alcohol precipitation. Many employ the same glass fiber membranes used for plasmid isolation, while others such as the Montage BAC96 Miniprep Kit (Millipore) use size exclusion membranes. A 96-well plate of cultures can be processed in approximately 60 min with these vacuum or centrifuge-based filtration systems. Typical yields range from 0.5 to 1 |g from a 1-ml culture.
RCA can be used to amplify large constructs giving a much higher yield of DNA than alkaline lysis-based methods. Approximately 5 |g of DNA can be obtained with TempliPhi Large Construct DNA Amplification kit (GE Healthcare) in 18 h from 1 ng of starting DNA. Random hexamers in the kit will amplify any DNA in the reaction so that higher levels of chromosomal DNA are often present in the amplification product if purified BAC DNA is not used as the starting material. As a result, and because of the large size of BAC clones, a higher concentration of DNA may have to be used in the sequencing reaction, and the RCA product is not ideal for library construction. The advantage of the method is that virtually any form of DNA can be the starting material, such as glycerol stocks or colonies, eliminating the need for culture growth.
9. Summary
The quality of sequencing template DNA directly affects the quality of sequence data obtainable. There are many template preparation methods available but no single method is perfect for all choices of vector, host strain, and sequencing application. Alkaline lysis remains the most popular method for isolating plasmid DNA but this and other inexpensive methods tend to be more time consuming, are difficult to automate, and suffer from low and variable yields. Consistent yield is the main concern when sequencing large numbers of templates, or when using a wide variety of vectors or hosts. It is difficult to establish a high-throughput sequencing pipeline when template yields are inconsistent. The column- or filter-based DNA purification methods offer higher yields and a higher purity product but still require many time-consuming steps and are subject to the same sample variability issues. These products are more expensive than simple miniprep methods. The SPRI technology eliminates many of the laborious steps of the traditional methods and as such is one of the quickest DNA purification methods available. Amplification technologies currently offer the most consistent yield and greatest flexibility for sequencing template preparation. RCA may be the method of choice when reliability, despite variation in vector and host strain, is of paramount importance. The RCA method eliminates culturing and purification steps, which can make it an attractive alternative despite higher initial costs. As DNA quality, quantity, and consistency vary between methods and with differences in host strain and vector, the choice of method has to be carefully considered, and more than one method may be required to meet all the sequencing template preparation needs.