1. Introduction
A physical map provides an ordering of clones, markers, or both. A physical map may be built using marker-clone associations (see Article 13, YAC-STS content mapping, Volume 3), where the markers are ordered such that they are contiguous for each clone (e.g., Alizadeh et al., 1995; Soderlund and Dunham, 1995). Alternatively, a physical map can be built using restriction fragment fingerprinting. In this case, a clone is digested with one or more restriction enzymes and the resulting fragments are measured. Two clones may overlap if they have a sufficient number of similar fragments. Overlapping clones are arranged into contigs to position the clones relative to each other. Whole-genome fingerprinting was first performed in the late 1980s (Coulson et al., 1986; Olson et al., 1986). Techniques for agarose-based fingerprinting have been greatly improved in order to reduce the amount of error (Marra etal., 1997). An alternative fingerprinting method called HICF (High Information Content Fingerprinting, Ding etal., 1999, 2001; Luo etal., 2003) has recently emerged. The most popular software for assembling fingerprinted clones into contigs is FPC (FingerPrinted Contigs, Soderlund et al., 1997), which works with either agarose or HICF. The FPC V7.2 software, executables, tutorial, and web-based tools are available from http://www.genome.arizona.edu/software/fpc.
2. The FPC software
FPC takes as input files of clones, where each clone is represented by a set of restriction fragments (often referred to as bands). It compares all pairs of clones, counts the number of shared bands, and computes the Sulston score (Sulston et al., 1988), which is the probability that the shared bands are just a coincidence. The user sets a cutoff and all clone pairs that have a Sulston score below the cutoff are considered overlapping. The assembly algorithm clusters clones such that each clone in a contig has a good overlap with at least one other clone in the contig. It then orders the clones by building a consensus band (CB) map, which is an approximation of the way the bands are ordered along the underlying genome. The clones are aligned to the CB map to give them an approximate position.
The measurement of the bands is not exact; so to compensate for this, the user supplies a tolerance to be used by FPC. If two bands are of the same value within plus/minus of the tolerance, they are considered to represent a fragment of the same size. False positive (F+) and false negative (F-) bands can cause F+ and F- clone overlaps; therefore, the accuracy of the band measurement is very important. It also affects the positioning of the clones; the more error in the data, the more imprecise the clone coordinates (Soderlund et al., 2000).
The user-supplied cutoff must be set to reduce F+ and F- overlaps. F+ overlaps result in chimeric contigs, which can generally be detected by an abundance of Q (Questionable) clones, where a Q clone is one in which the ordering routine cannot align 50% or more of the bands to the CB map. F- overlaps cause the clones to assemble into many contigs. For example, given a cutoff that results in 70% overlap between clones (which is typical for agarose-based fingerprints), a genome size of 2400 Mb, clones of size 150 000, and a 17x coverage, the clones will assemble into 1574 contigs if the clones are evenly distributed (Lander and Waterman, 1988). Since some regions are not cloneable and the coverage of clones is not evenly distributed, the number of contigs will be much greater than 1574.
The main FPC automatic functions are: (1) Build contigs, (2) IBC (Incremental Build Contigs) adds new clones to existing contigs and merges contigs, (3) DQer reassembles contigs with over a given number of Q clones using a more stringent cutoff, which reduces F+ overlaps, and (4) End Merger compares clones at the end of contigs using a less stringent cutoff and automatically joins contigs (V7.2 only), which reduces F- overlaps. As these functions do not fix all F+ and F-overlaps, FPC also contains many interactive queries and edit functions so that the user can manually fix the remaining problems (Engler and Soderlund, 2002).
2.1. Using markers and anchors in FPC
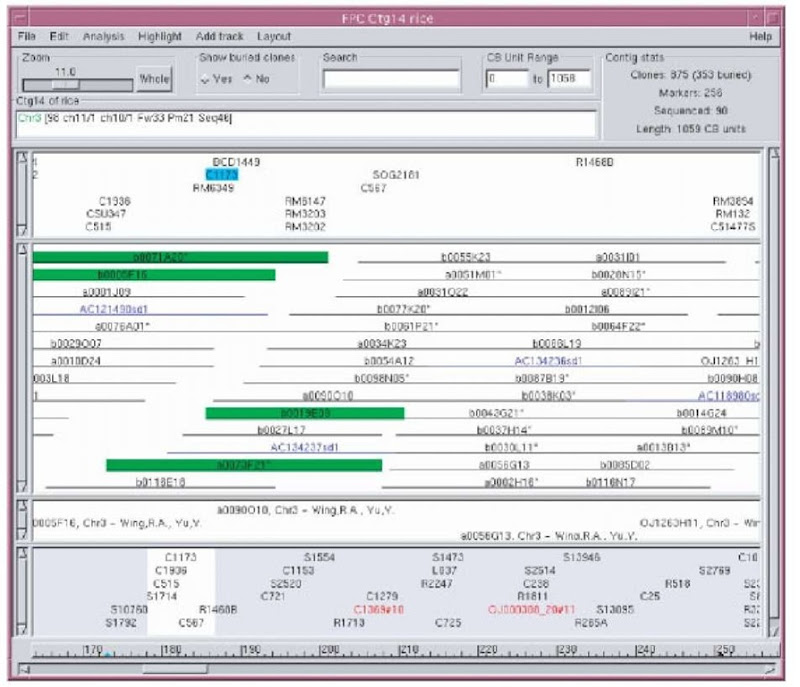
Fingerprints can be assembled to order the clones relative to each other, but do not order contigs or position them on the chromosome. Genetic markers or radiation hybrid markers (see Article 14, The construction and use of radiation hybrid maps in genomic research, Volume 3) have order and location on the chromosomes. If these markers have been hybridized to fingerprinted clones, the data can be entered into FPC and used to anchor contigs to chromosomes. Unanchored markers, such as many of the ESTs, are often hybridized against the clones. These marker-clone associations can be entered into FPC, which gives the markers an approximate ordering. The presence of markers in the FPC map is also important for verifying fingerprint data and can be used in conjunction with the fingerprints for assembly. The contig display (see Figure 1) provides a versatile way of viewing the clones, markers, and anchors.
When BESs (BAC end sequences) or sequenced clones (draft or finished) are associated with clones in the map, additional sequenced markers can be added electronically. This is done using the FPC function BSS (Blast Some Sequence), which takes a file of markers, compares them against the sequences associated with FPC clones using BLAST (Altschul et al., 1997), megaBLAST (Zhang et al., 2000), or BLAT (Kent, 2002). The hits can be added to the FPC map as electronic markers.
Figure 1 Each of the four regions with a scroll bar on the left is referred to as a track. The first track shows the markers. Selecting a marker highlights the clones that it is contained in, as illustrated by marker C1173. The second track shows the clones. The blue clones starting with “A” are sequenced clones from Genbank that have been digested in silico using FSD (FPC Simulated Digest, Engler et al., 2003). The third track shows remarks associated with clones or markers. The remarks shown here are attached to the simulated digest clones. The lowest track shows all anchors, which are markers that have a chromosome position. Anchors shown in red disagree with the majority of anchors as to the chromosome assignment. The chromosome assignment is shown above the first track and has been assigned by an FPC function based on majority rules
2.2. Sequencing
FPC is used for selecting clones for sequencing (e.g., The International Human Genome Mapping Consortium, 2001). Until recently, this has been performed interactively with FPC tools. A recent release provides a routine that automatically selects an MTP (Minimal Tiling Path, Engler etal., 2003), using sequence similarity or fingerprint overlap. When a draft sequence hits two BESs of clones that are near each other in FPC, this dual information provides a reliable overlap known in bases (e.g., Chen et al., 2004). For finding overlaps based on fingerprints, the algorithm looks for overlapping clones that are confirmed by two flanking clones and one spanner. An MTP is selected from the overlapping pairs using Dikstra’s shortest path algorithm (Dijkstra, 1959), giving precedence to sequence-based overlaps.
2.3. Agarose versus HICF
A commonly used implementation of agarose-based fingerprinting uses one 6-base enzyme and produces fragments with an average size of 4096 bases, which results in approximately 30-50 bands per clone. Typically, the program Image (Sulston etal., 1989) is used to determine the migration rate of the fragments and the corresponding sizes of each band. The accumulative size of fragments is used as the approximate size of the clone. A bottleneck with this method is the human time spent in interactively calling the bands in Image; this problem has recently been resolved with BandLeader (Fuhrmann et al., 2003).
HICF uses multiple enzymes and detects the terminal base of each fragment. Hence, two bands are considered the same if they have the same size and terminal base pair. The bands are run on a sequencing machine so that we have high-precision measurements of the bands. The bands sizes range from 50 to 500, and clones typically have over 100 bands; note that the bands only cover a subset of the clone, so they cannot be used to calculate the approximate size of the clone. Though FPC does not take base information as input, Ding et al. (1999) developed a simple scheme to encode the base in the fragment size.