1. Introduction
Identifying genomic regions involved in the adaptive divergence of closely related species or populations is a major goal of evolutionary genetics research. Adaptations come about through a variety of modes, including global selection for a particular allele, local adaptation, where alternative alleles are favored in different environments, and balancing selection (heterozygote advantage), where increased diversity is favored at a particular locus. These modes of selection make distinct predictions regarding the distribution of inter- and intraspecific genetic variation near the selected region, thus leaving a distinguishing “signature”. For example, repeated favorable changes to a protein lead to accelerated rates of amino acid evolution between species relative to synonymous substitution rates, while long-term heterozygote advantage increases intraspecies diversity around the selected site (Hudson and Kaplan 1988).
2. Using interspecies divergence
Over the past three decades, mathematical and computer models have helped to characterize the signature of natural selection on nucleotide sequence variation (e.g., Maynard Smith and Haigh 1974; Hudson and Kaplan 1988; Takahata 1990). On the basis of these characterizations, statistical approaches have been developed to identify genomic targets of adaptation and make inferences about current and past selective pressures. An important class of methods aims to detect nonneutral evolution of amino acid sequences using phylogenetic patterns of synonymous and nonsynonymous variation (for a review, see Yang and Bielawski 2000). These approaches have been successful in identifying a subset of proteins with an accelerated rate of amino acid evolution (e.g., Clark et al., 2003), but cannot detect adaptations consisting of few substitutions to a given protein or changes that occur outside of the coding region.
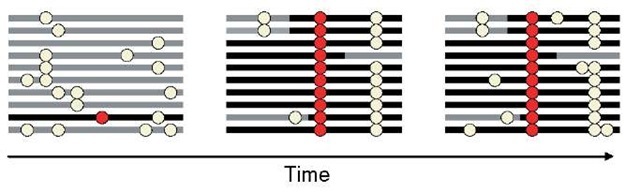
Figure 1 The effect of a “selective sweep” on linked neutral variability. Each line represents a chromosome and the circles the nonancestral allele at a site. Light yellow circles are mutations with no fitness effect, i.e., which evolve neutrally. A favorable mutation arises (shown in red) and, because it is favored, increases rapidly in frequency in the population. It recombines onto a limited number of haplotypes during its ascent; how much recombination occurs depends on the recombination rate and the strength of selection. After fixation, variability is replenished by mutation. However, for a while, most mutations are new and therefore tend to be at low frequency
3. Using polymorphism data
Regulatory adaptations or subtle adaptations in proteins may be detectable from patterns of polymorphism in samples of extant individuals, so long as the selective events are recent. Indeed, when a rare allele arises and rapidly fixes in the population, it distorts patterns of variation at linked neutral sites relative to the expectation in the absence of natural selection (Maynard Smith and Haigh 1974) (see Figure 1). Under simplifying assumptions, this signature of a “selective sweep” can be exploited to find regions that have been the target of positive selection in the past ~N e generations, where N e is the diploid “effective population size” of the species (Przeworski 2002). In humans, estimates of Ne are on the order of 10 000, suggesting that polymorphism-based approaches can be informative about selective events over the past ~250 000 years, including those associated with the emergence of anatomically modern humans.
The most common methods used to identify adaptive genetic changes from polymorphism data are the so-called tests of neutrality. These tests begin by assuming a neutral null model of a random-mating population of constant size (the “standard neutral model”) and then assess whether the value of some summary of the data