Introduction
Hair is commonly encountered as physical evidence in a wide variety of crimes. Human hair is understandably of special importance in criminal cases involving personal contact, such as homicide and sexual assault. In some cases, animal hair can also be important.
Since hair continually falls from the body of every person it is often present at the crime scene or on the clothing of the victim and suspect. In general, hair is not easily destroyed except for its decomposition by microbial or insect invasion, and even after soft tissue decomposition, hair remains useful for personal identification and comparison for a long period of time.
Any review of the forensic aspects of hair examination must start with the observation that it is not yet possible to individualize a human hair to any single head or body. This does not imply that hair has no value as physical evidence. At present the initial method used for forensic hair examination is microscopical comparison of questioned hairs collected from the crime scene to standard hairs of the victim and suspect. In general, 25 standard hairs from at least five locations of the scalp would be collected from all individuals involved in the case as representative samples of an individual, since most head hairs are variable and of different color shade. In some laboratories, additional examinations such as ABO blood typing, elemental analysis and electro-phoretic analysis of hair proteins have been carried out for characterizing hair samples. When hairs are properly collected at the crime scene and their submission to the laboratory is accompanied by an adequate number of control hairs, and the comprehensive examinations described above are performed, hair can provide strong corroborative evidence for placing an individual at a crime site. In order to enhance the degree of certainty of identification, DNA typing has been applied to forensic hair comparison.
Nuclear DNA in Hair
DNA is housed within the nucleus of all types of cells in the human body, except mature red blood cells. Hair is an appendage of the skin that grows out of a histological organ known as the hair follicle and naturally contains DNA. Hair growth stage is an important factor for extracting DNA from hair.
Hair grows cyclically with alternating periods of growth and quiescence. The hair follicle has three distinct growth phases. During growth, the follicle is in the anagen phase (Fig. 1). In the anagen phase, mitotically active cells above and around the dermal papilla of the follicle grow upward to form the medulla, cortex, cuticle and inner root sheath of hair. The subsequent resting state is called the telogen phase (Fig. 2). In the telogen phase, the hair is anchored in the follicle by only a club. The transition period, when follicle is moving into an inactive quasi-embryonal state, is called the catagen phase. On a healthy head hair, one would expect to find 80-90% of the hair follicles are at the anagen phase of the growth cycle, 2% at the catagen phase, and 10-18% at the telogen phase. If hairs were forcibly pulled from the head of a criminal during a struggle and remained at the crime scene, hair samples as physical evidence would be in the anagen phase (plucked hair), containing many cells of the inner root sheath. From such plucked hairs, it is easy to extract the DNA by the same procedure as used for tissue samples because a greater part of DNA in hair is located in the root and surrounding sheath cells. A single plucked hair may contain as much as 100-500 ng of DNA. However, in most crime cases, human hair samples recovered from the scene of a crime are in the telogen phase (shed hair) which has naturally fallen out. In a hit-and-run case, only the hair shaft without the root region may be collected from a suspect car and submitted to the laboratory as physical evidence. In such a case, the DNA would have to be extracted from the nuclear remnant of the club and hair shaft (Fig. 3).

Figure 1 An anagen human hair root forcibly pulled from the scalp.

Figure 2 The telogen root of a human hair which fell from the scalp.
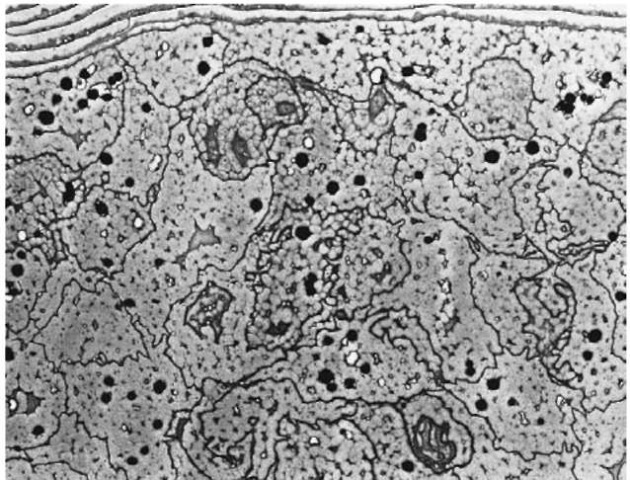
Figure 3 Transmission electron micrograph of human head hair. Each cortical cell contains nuclear remnants and micro fibrils. Melanin granules are scattered throughout the cortex.
DNA extraction from hair
The generic procedure for DNA isolation from evidence samples is to digest the sample with a TNE buffer (10 mM Tris-HCl, 100 mM NaCl and 10 mM EDTA, pH 8.0) containing proteinase K followed by phenol/chloroform extraction. It is difficult to isolate DNA from shed hair by routine procedures because the shed hair contains very small and extremely degraded DNA. In order to effectively extract DNA from the shaft and root of this type of hair, keratinized cells of the hair must be completely broken down by the digestion buffer. Although the standard TNE buffer leads to partial dissolution of the hair shaft, a two-step digestion method using TNE buffer facilitates its dissolution. A digestion buffer containing calcium as activator of proteinase K also leads to total dissolution of the hair.
The DNA in the phenol/chloroform extract must be separated from the extraction solvents and concentrated prior to DNA amplification. For concentration and purification of DNA, several techniques such as the CTAB (cetyltrimethylammonium bromide) treatment, the GENECLENE (Bio 101) and the Microcon (Amicon) have been tested. In general, the effective concentration of DNA is performed by using spin ultrafiltration concentrators such as the Microcon equipment.
The procedure of DNA extraction from the shaft or root of shed hair is as follows.
A single shed hair is washed in a detergent (1% Tween-20), deionized sterilized water and absolute ethanol. The hair sample is cut at the level of 5 mm from the root and divided into the shaft and root. Five cm from the proximal end of the hair shaft is used for DNA extraction. The hair shaft is cut into 5-10 mm pieces with a sterilized blade, and placed in a 1.5 ml microcentrifuge tube and then incubated in a digestion buffer (320 ul of TNE buffer, 40 ul of 10% SDS (sodium dodecyl sulfate), 40 ul of 0.4 M DTT (dithio-threitol) and 15 ulof10mgml_1 proteinase K) at 55°C for 1 h. Then a second incubation is performed at 55°C for 1 h after addition of 20 ul of 10% SDS, 20 ul of 0.4 M DTT and 5 ulof10mgml-1 proteinase K.
The DNA solution is then washed three times with TE-saturated (10 mM Tris-HCl and 1 mM EDTA) phenol (pH 8.0), once with phenol chloroform, once with chloroform, and once with ether. The DNA in the aqueous phase is then purified and concentrated using Microcon 100 ultrafiltration. Extraction of DNA from the hair root (3 mm in length) is performed by the same method.
In some reports, the amount of DNA in the hair shaft usually cannot be amplified to detectable levels with 30-35 cycles of PCR (polymerase chain reaction). However, according to one study, the DNA extracted from a 5 cm length of head hair shaft was about 20 ng and its base pair size was demonstrated to be lower than 500 bp (base pairs), mainly 100 bp by agarose gel electrophoresis.
Amplification and typing of products
PCR amplification PCR is a process by which short segments of DNA sequence can be selectively replicated a millionfold or more. The PCR process consists of three major steps: denaturing, annealing and extension. The denaturing step is the separation of the two strands of the DNA molecule. The double-stranded DNA is dissociated into single strands by incubation at 94-96°C. The two strands serve as templates for the replication of their complementary sequences. Primer binding is called annealing. The temperature is lowered (55-72°C) to allow the oligonucleotide replication primers to bind to their complementary sequences on the template DNA strands. In the next step (72-75°C), the primers are extended by the stepwise addition of nucleotides to the 3′ end of the primer strand using the target sequence as template. This step is mediated by DNA polymerase.
For hair shaft samples, 5 ul of DNA solution (corresponding to 2.5 cm of hair shaft length) is used as a template DNA. It has been reported that melanin contained in the hair is often extracted together with DNA and acts as an inhibitor of PCR amplification (Fig. 4). When melanin is present at concentrations higher than 1.6 ng ul-1 in the PCR solution, PCR amplification was significantly inhibited (Fig. 4). However, most melanin is preferentially removed from the extract by the phenol/chloroform extraction procedure.
DNA typing A number of DNA typing systems after the PCR amplification are available: AmpliFLP (amplified fragment length polymorphism); STRs (short tandem repeats); HLA DQ a and Polymarker (PM). In AmpliFLP analysis (D1S80, D17S30 etc.), the template DNA is amplified using primers which flank a core repeat of 8-16 bp. Fragment length polymorphism, like VNTR (variable number tandem repeat) polymorphism, is based on the number of core repeats found in a particular allele. AmpFLP alleles range in size from 100 to 1300 bp, whereas VNTR alleles range from 500 to 23 000 bp. STRs are similar to the AmpFLP system, except that the core repeat is shorter. STR alleles range from 100 to 400 bp and their core repeats are 4-6 bp. In HLA DQ a and PM, the amplified DNA products are typed by dot-blot analysis. The probes detect six common DQ a alleles which, in combination, determine 21 possible genotypes. PM consists of five loci, low density lipoprotein receptor (LDLR, 214 bp), glycophorin A (GYPA, 190 bp), hemoglobin gammaglobin (HBGG, 172 bp), D7S8 (151 bp) and group specific component (GC, 138 bp). PM typing has been developed in which several markers at different loci are analyzed at the same time by the same technique used in HLA DQ a analysis.
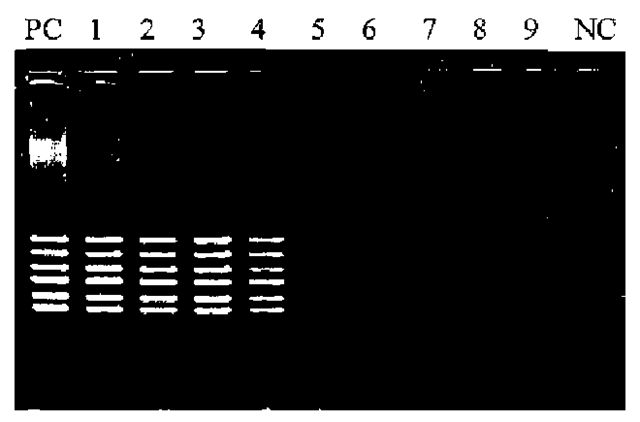
Figure 4 Influence of melanin on PCR of PM kit. PC: Positive control (melanin free); 1: 0.1 ng ul-1 melanin in PCR solution; 2: 0.2ng ul-1; 3: 0.4ng ul-1; 4: 0.8ng ul-1; 5: 1.6ng ul-1; 6: 3.2ng ul-1, 7: 6.4ng ul-1; 8: 12.8ng ul-1; 9: 25.6ng ul-1; NC: negative control.
Because the size of the DNA extracted from shed hair may be 100-500 bp, the loci with a small amplification size have to be used as a target for DNA typing of this type of hair. For this reason, it seems that STRs and PM typing are appropriate methods for routine hair comparison work.
There are approximately 4.0 x 108 STR loci dispersed throughout the human genome; SRT loci, TH01, CSF1PO, VWA, FGA, TPOX etc. are used for biological evidence. The amplification product is subjected to a polyacrylamide gel electrophoresis and then visualized by silver staining. A number of STR loci may be amplified and separated simultaneously, a technique known as multiplexing. Multiplex PCR kits such as Blue kit (FGA, vWA, D3S1358), Green kit (CSF1PO, TPOX, TH01) and Profiler (FGA, vWA, D3S1358, CSF1PO, TPOX, TH01, D7S820, D13S317, D5S818, PE/Applied Biosystems) are currently commercially available. The amplification products are electrophoresed and analyzed on a fragment analyzer. The combination use of STR loci enormously enhances the discrimination power of individuals, whereas in the severe degraded samples some loci of the multiplex system cannot be detected. For typing of DNA extracted from the hair, a single locus amplification may be recommended. TH01 and PM typing for the shed hairs are shown in Figs 5 and 6, respectively. TH01 locus was amplified using Quick-Type® HUMTH01 (Lifecords). PM typing was performed using AmpliType PM PCR Amplification and Typing kit (Perkin-Elmer). In PM typing, it is generally recommended that a DNA probe strip with no visible ‘S’ dot should not be typed for any locus (‘S’ dot is a minimum dot intensity control and corresponds to the ‘C’ dot on the HLA DQ a DNA probe strip). However, by comparing all bands of amplified DNA sample with relevant bands of amplified control DNA (template: 2 ng, provided in the kit) by agarose gel electrophoresis, all five or a few loci could be correctly typed even when the ‘S’ dot was not visible on the DNA probe strip. The success rate for DNA typing of the shaft and root of shed hair is estimated at about 25% and 50%, respectively. This suggests that the hair root is more suitable for STRs and PM typing when shed hair is the evidential sample. However, even in the root of shed hair, about 50% of samples cannot be analyzed by these DNA typing systems because of its limited and degraded nuclear DNA.
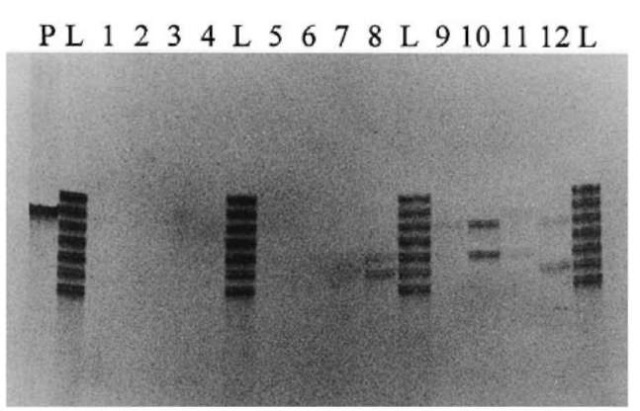
Figure 5 TH01 typing from head hair shaft of the telogen phase on 4% denatured polyacrylamide gel electrophoresis. P: positive control (K562 DNA); L: allelic ladder; 1-12: subjects 112; 8: 6-7; 10: 7-9; 12: 6-9.
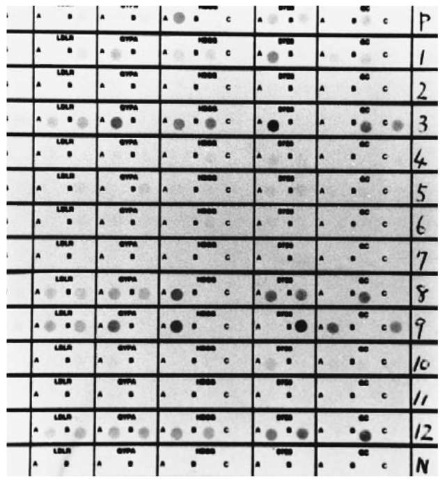
Figure 6 PM typing from head hair root of the telogen phase according to the protocol in the PM kit. P: positive control (K562 DNA); 1-12: subjects 1-12; N: negative control, 1: LDLR; BB, GYPA; AA, HBGG; AB, D7S8; AA, GC; AB, 3: AB, AA, AB, AA, BC, 8: AB, AB, AA, AB, BB, 9: AB, AA, AA, BB, AC, 12: AB, AB,AB, AB, BB.
Mitochondrial DNA
In addition to nuclear DNA, eukaryotic cells contain a unique mitochondrial form of DNA. The inheritance of mitochondrial genes is exclusively maternal, and clearly non-Mendelian. Mitochondria in human cells contain between 2 and 10 copies of a double-stranded, circular DNA molecule that is about 16.5 kb in length. Owing to the high copy number of mitochondrial DNA molecules per mitochondrion and the high number of mitochondria per cell, there is 1000-10 000-fold molar excess of mitochondrial DNA (mtDNA) compared to nuclear DNA.
Hypervariable regions
The mitochondrial genome contains a noncoding hypervariable control region. In particular, there are two segments, hypervariable region 1 (HV1:16024-16365) and hypervariable region 2 (HV2:73-340) within this 1100 bp control region (Fig. 7), also called the D-loop, that tend to mutate with an extremely high frequency, at least 5-10 times that of nuclear DNA. This high copy number and the high mutation rate of mtDNA are the main reasons for its use in individual identification. The complete 16 569 nucleotide sequence of human mtDNA has been established for a reference individual. For present, all comparisons are made to this reference called the Anderson sequence.
Amplification and sequencing of mtDNA
PCR amplification In general, HV1 and HV2 in control region are subjected to mtDNA analysis. Several protocols for the PCR amplification of mtDNA control region and sequence analysis of the PCR product have been established.
Two sets of PCR primers are used to amplify overlapping segments from both HV1 and HV2. Examples of PCR primers used for the hair sample are shown in Table 1. The primer pairs used to amplify HV1 and HV2 are: HV1-A; A1/B2, HV1-B; A2/B1,HV2-A; C1/D2, HV2-B; C2/D1.
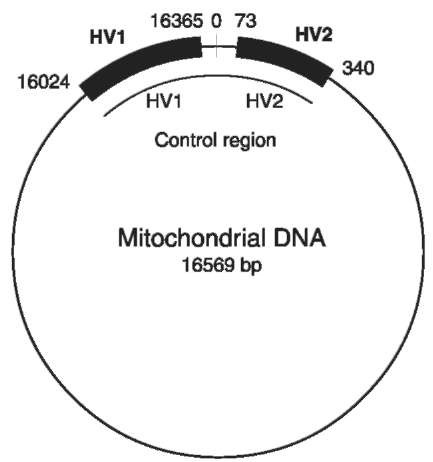
Figure 7 Locations of the two HV regions on the mtDNA circle.
Amplification is performed in a reaction mixture of 25 ul using a thermal cycler. Each reaction mixture comprises 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 15 mM MgCl2, 10.0 pmol of each primer (0.2 uM), 200 uM each dNTP, 2.5 U Taqpolymerase (Ampli-TaqGold, Perkin-Elmer) and 5 ul of the hair extract DNA (corresponding to 5 mm of hair shaft length). Thermal cycling conditions are 9 min at 95°C, followed by 36 cycles of 95°C for 10 s, 54°C for 30 s and 72°C for 30 s. A portion of the PCR products are taken to verify the amplification of the expected fragment by agarose gel electrophoresis and ethidium bromide staining prior to DNA sequencing (Fig. 8).
DNA sequence DNA sequencing is performed on mtDNA PCR products using fluorescence-based chemistry. In general, two types of fluorescent dye methods, namely a dye terminator cycle sequencing and dye primer cycle sequencing are used. The PCR products must be purified prior to DNA sequencing to remove unincorporated PCR primers and excessive dNTPs. The purified PCR products are quantified using a fluorometer, and diluted with deionized water to a suitable concentration. For example, sequencing fragments are generated using the dye terminator cycle sequencing kit (Applied Biosystems). Following gel filtration, the purified extension products are dried in a vacuum centrifuge. The purified products are electrophoresed on a 5-6% polyacryla-mide sequencing gel on an AB 373A DNA sequencer or 377 DNA sequencer and the AB computer software collects and analyzes the raw data.
Table 1 Examples of primer set for hair samples
| Primer | Forward primer | Reverse primer |
| set | light strand | heavy strand |
| HV1-A | L15997 | H16236 |
| 5′-CACCATTAGCACCCAAAGCT-3′ | 5′-CTTTGGAGTTGCAGTTGATG-3′ | |
| HV1-B | L16159 | H16401 |
| 5′-TACTTGACCACCTGTAGTAC-3′ | 5′-TGATTTCACGGAGGATGGTG-3′ | |
| HV2-A | L048 | H285 |
| 5′-CTCACGGGAGCTCTCCATGC-3′ | 5′-GGGGTTTGGTGGAAAIIIIIIIG-3′ | |
| HV2-B | L172 | H408 |
| 5′-ATTATTTATCGCACCTACGT-3′ | 5′-CTGTTAAAAGTGCATACCGCCA-3′ |
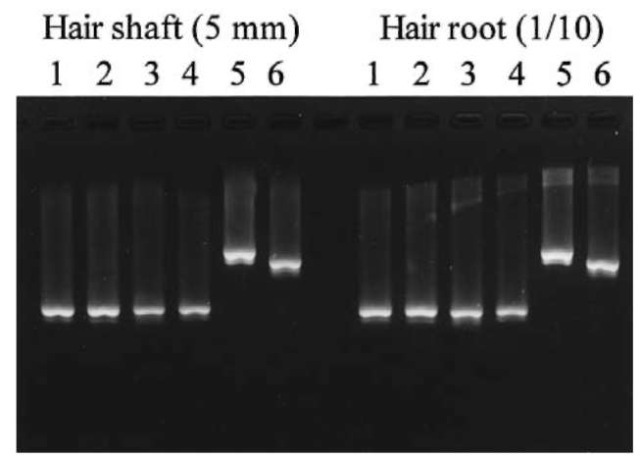
Figure 8 Agarose gel electrophoresis of PCR of mtDNA extracted from head telogen hair. Template DNA corresponds to the amount of DNA extracted from a 5mm length of the hair shaft and one-tenth of the hair root. Primer pair: 1; A1/B2, 2; A2/B1, 3; C1/D2, 4; C2/D1, 5; A1/B1, 6; C1/D1.
Sequence data interpretation When the sequencing is successfully achieved from both forward and reverse direction (Fig. 9), the sequences are aligned and compared with the reference sequence using the Sequence Navigator computer program (Applied Bio-systems). Each nucleotide substitution, deletion or insertion is registered. Polymorphism of mtDNA in an individual is determined based on the difference of nucleotides at some position between the sequence obtained from the individual and the reference sequence (Fig. 10). However, the sequences cannot be determined in some individuals who have an anomaly called heteroplasmy within the mtDNA control region. There are two kinds of heteroplasmy in human mtDNA; sequence heteroplasmy and length hetero-plasmy. Sequence heteroplasmy is a condition in which more than a single base is observed as mixed peaks at a particular position in electropherogram. This indicates the presence of two variant populations of mtDNA in the same individual. In length heteroplasmy, because of the existence of more than two variants in length in a homopolymeric tract of cytosines between nucleotides 16184 and 16193, the sequences of 3′-side lying ahead of this tract are hardly determined. Because the presence of hetero-plasmy might potentially lead to a false exclusion in a forensic case, careful analysis and comparison of known and questioned samples is necessary.
Currently the mtDNA is being used forensically, and its success rate for analysis of questioned samples is over 80%. At the present, mtDNA analysis is the most effective method for hair sample to which nuclear DNA typing cannot be applied.
Figure 9 Electropherogram for the forward HV1-A (Primer A1) analyzed byABI PRISM.

Figure 10 A comparison between hair DNA obtained from five individuals and reference sequence. Pyrimidine substitutions (C-Tor T-C transition) are found at 16243 (D), 16249 (B), 16261 (A and C), 16266 (E) and 16278 (D and E). C-A and C-G trans-versions are observed at 16257 (C) and 16266 (A), respectively.
Factors Causing DNA Damage Environmental insults
Hair samples collected from the scene of a crime may have been exposed to environmental conditions such as lighting, high temperature and humidity, dust, soil, bacteria and so on. These various environmental exposures have an effect on DNA analysis.
A major consequence of ultraviolet (UV) damage caused by lighting is the formation of covalent links between adjacent pyrimidines to make pyrimidine dimers. PCR product yields are reduced as the dose of UV exposure is increased because Taq polymerase stops at UV damage sites. If hair samples are exposed to lighting for a long period and received very high UV doses, PCR is completely inhibited. Microbial contamination occurs when hair samples are located in the dust or soil under the warm and moist conditions. In this case, degradation of DNA is caused by microbial activity which causes strand scission and results in the preferential loss of the larger allele products.
Cosmetic treatments
Hair color is altered by bleaching and dyeing, and form is changed by styling such as permanent waving. Any of these procedures may produce irreversible damage to the hair shaft.
Hair bleaching using hydrogen peroxide at alkaline pH selectively decolorizes the natural hair pigment. Permanent hair dye is a system of hair coloring in which the cortex of the hair is colored by reaction of the colorless dye precursors with hydrogen peroxide. In both hair bleaching and coloring, oxidation processes are used to break down melanin granules. Such oxidizing agents cause degradation of DNA in the hair shaft, such as base modification and strand scission.
In permanent waving, three stages are involved; physical and chemical softening of the hair, reshaping and hardening of fibres to retain the reshaped position. A permanent curl in the hair is accomplished by breaking the disulfide linkage of keratin with a reducing agent and reforming the pliable hair with a curling rod. The hair is then oxidized with agents such as hydrogen peroxide, sodium perborate or sodium percarbonate, so that the disulfide bonds are reconstructed and the curl fixed. It is generally accepted that organic solvents and reducing agents have no effect on the quality of DNA. However, the DNA in the hair shaft is damaged by oxidation in the last stage of permanent waving.
