Introduction
The development of methods for the amplification and detection of DNA fragments using the polymerase chain reaction (PCR) has resulted in rapid and dramatic advances in forensic DNA typing. Using the PCR it is possible to easily produce analytically significant amounts of a specified DNA product from trace quantities of DNA. In its forensic application, the PCR is used to demarcate and amplify known polymorphic sites on a distinct chromosome and produce discrete and easily characterized fragments of DNA.
The introduction of PCR-based forensic assays has also resulted in a need for efficient and automated procedures for analysis of the reaction products. This requirement has been the driving force behind the development of capillary electrophoresis (CE) methods for DNA analysis. In CE, DNA separations are performed in a thin 50-75 um fused silica capillary filled with a sieving buffer. These capillaries have excellent capabilities to dissipate heat, permitting high electric field strengths to be used. As a result separations in capillaries are rapid and efficient.
Additionally, the capillary can be easily manipulated for efficient and automated injections. Detection occurs via adsorption or fluorescence through a window etched in the capillary.
When used in short tandem repeat (STR) analysis, CE systems require specialized techniques. The high ionic strength of the PCR reaction mixture inhibits CE injection methods, and separations must be performed using highly viscous buffers. Detection of STRs is generally carried out by laser-induced fluorescence as adsorption techniques have poor sensitivity. Lastly, the serial nature of CE separations requires internal and external standardization to achieve highly precise measurements.
Theory of CE Separations
DNA fragments can be difficult to separate under normal CE conditions due to their virtually constant charge to mass ratio. Therefore, analyses are performed using a replaceable sieving matrix consisting of a water-soluble polymer dissolved in a suitable buffer. Such solutions are referred to as ‘physical gels’ as they are not chemically crosslinked. This fact makes them different from the crosslinked or ‘chemical gels’ used in slabgel analysis. The advantage of physical gels is that fresh gel solution can be pumped into the capillary at the conclusion of each analysis, thus limiting problems with carryover. Experiments carried out using a variety of physical gels have shown that with careful optimization of molecular weight and concentration, high resolution DNA separations can be produced.
Several different mechanisms have been postulated to describe the separation of DNA in physical gels. These include transient entanglement coupling, Ogston sieving, and reptation. At low concentrations of polymer, separation takes place by means of a frictional interaction between the DNA and the polymer strands. This mechanism is known as transient entanglement coupling. At higher concentrations of polymer, individual polymer molecule strands begin to interact, producing a mesh. The polymer concentration at which this occurs is known as the entanglement threshold. Above the entanglement threshold, DNA fragments separate by sieving through transient pores created in the polymer mesh (Fig. 1). Fragments which are larger than the average pore size reptate or move in a snakelike manner through the mesh. The key to producing an acceptable separation is to specify a polymer concentration at which the size of these virtual pores approximates the radius of gyration of the DNA in solution.
To keep solution viscosity manageable, the polymer length must be kept to a minimum. Other characteristics of importance include the relative stiffness and polydispersity of the polymer. The key to producing an acceptable separation is to optimize the polymer molecular weight and concentration in the buffer solution using the resolution and mobility of the DNA as a guide to system performance. Other parameters such as column temperature, and applied electrophoresis voltage must also be tested. These variables affect resolution by affecting the rate of diffusion of the DNA as it moves down the capillary.
In uncoated capillary columns, charged silanol groups on the silica surface induce buffer ions to form a double layer along the capillary walls. Application of the electric field induces a flow of the bulk solution toward the negative electrode. This effect is known as electroosmotic flow (EOF). The magnitude and direction of the EOF is dependent on the number and type of active sites on the capillary surface and the pH of buffer. Electroosmotic flow can be considered a detriment to stable DNA separations because its velocity can change with each run, making peak migration times vary.
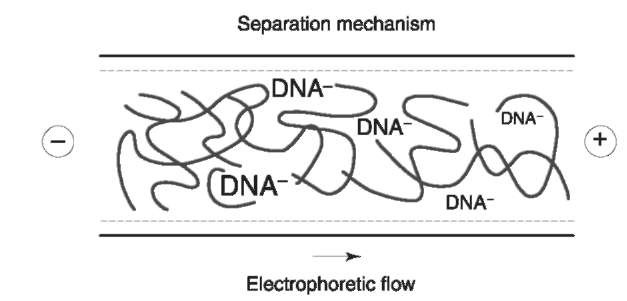
Figure 1 DNA is sieved through transient pores created in the polymer mesh. Smaller fragments are less impeded by the mesh and elute first.
To control EOF, capillary columns must be coated to mask these charged sites. Coatings can be classified into two types: dynamic coatings that must be periodically replenished or static coatings that are bonded to the capillary walls. Dynamic coatings are compounds added to the buffer, which mask active sites on the capillary walls. Many of the polymers developed for DNA separations perform this function, dynamically coating the capillary as well as sieving the DNA. Capillaries may also be washed with dilute HCl just prior to filling with buffer, to neutralize active sites on the capillary walls.
Static coatings are inert substances that are covalently bound to the internal walls of the capillary. Such coatings must be stable at the pH of analysis and free from contamination. The key factor in selecting a coating is its stability and lifetime under the conditions at which it is used. With periodic rinsing coated capillaries can last for months before failure.
Injection and Sample Preparation
There are two modes of injection in CE systems: hydrodynamic and electrokinetic. Hydrodynamic injections are performed using pressure to drive the samples into the capillary orifice. The solution viscosity, the capillary radius, and the applied pressure determine the volume of sample injected by this technique:
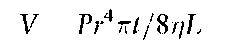
where P is the pressure, r is the radius of the capillary, t is the time and r\ is the viscosity. Hydrodynamic injections are particularly well suited for quantitative analyses of DNA. When properly initiated, highly reproducible quantities of sample may be introduced onto the capillary. Standard deviations of DNA peak area using this technique have been shown to be 3%. However, the wide injection bands produced tend to limit resolution.
Electrokinetic injections are performed using an applied voltage to induce the sample to migrate into the capillary orifice. The quantity of material injected QDNA onto the capillary by this technique can be described by the following
![]()
where E is the field strength, r is the capillary radius, u.ep is the electrophoretic flow and u.e of is the electro-osmotic flow. However, this equation must be modified as other ions present in solution will compete with DNA for the role as electrophoretic charge carriers. Thus, the quantity of DNA injected is also
a function of the ionic strength of the solution, and the total quantity injected:
![tmp4514_thumb[2] tmp4514_thumb[2]](http://lh5.ggpht.com/_1wtadqGaaPs/TFEuoBcHakI/AAAAAAAALPc/KEAFhhKtsV0/tmp4514_thumb2_thumb.png?imgmax=800)
Electrokinetic injections produce narrow injection zones but are highly sensitive to the sample matrix. For example, the electrokinetic injection of PCR products into a capillary is inhibited by the high salt content of the sample matrix (>50 mM Cl_). These small ions also tend to be selectively injected into the capillary. To overcome this problem, PCR samples can be purified by means of dialysis, spin columns or ethanol precipitation. The dialysis step appears to be the most effective for removing excess salt, whereas the spin columns are more effective at removing primer peaks, enzyme and dNTPs. Dilution of the sample in water or deionized formamide is another technique for reducing ionic strength. The DNA signal can then be selectively enhanced utilizing fluorescence detection.
The above steps increase quantity of sample injected by removing interferences or by sharpening the injection zone in a process known as stacking. Stacking, also called field amplified injection, occurs when the ionic strength of the sample zone is lower than that of the buffer. As the current through the system is constant, the lack of charge carriers in the sample zone produces a strong electric field that ends abruptly at the interface between the sample zone and the buffer inside the capillary (Fig. 2). DNA molecules mobilized by this field move rapidly towards the capillary as the injection voltage is applied and ‘stack’ in a narrow zone at the interface. Stacking allows a large sample zone to be loaded onto the capillary with a minimum of band broadening. Stacking also aids in producing efficient separations. With sharp injection zones, shorter capillaries and less gel media is required to effect a separation.
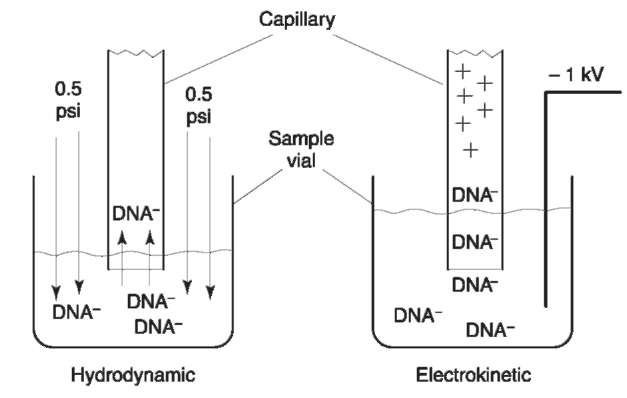
Figure 2 The two modes for injection for CE. In hydrodynamic injections, pressure is used to force sample into the capillary. In electrokinetic injections, an applied voltage causes the DNA to migrate into the capillary. Figure courtesy of Dr Butler, Gene-Trace Systems.
Detection and Data Analysis
Early capillary electrophoretic separations of PCR products utilized UV absorbance detection, and required extensive purification and concentration of the samples prior to analysis. The relatively short path length and the dispersion produced by the capillary walls limited sensitivity. Laser-induced fluorescence (LIF) solved this problem by focusing an intense beam of light directly onto the sample volume within the capillary. Detection enhancements of 400-fold or more have been achieved using LIF as compared to UV detection.
Fluorescence detection of DNA is achieved by derivatizing the DNA using dyes to produce fluorescent adducts. Native DNA fragments can be detected using fluorescent intercalating dyes. These dyes bind to the DNA molecule by binding to the DNA helix, affecting the configuration of the aromatic rings of the dye and producing a large enhancement of the fluorescence signal. Additionally, intercalating dyes help to minimize effects of DNA structure on migration rate, resulting in better estimates of fragment lengths. The low background fluorescence of the uncomplexed dyes allows them to be added directly to the CE buffer.
Dye molecules may also be covalently bound to the DNA fragments. This procedure is commonly used to label single-stranded DNA by attaching a dye molecule to the 5′ end of each primer. Typically, only one primer is labeled with a fluorescent dye to avoid doublet bands. Upon completion of the PCR, all of the target DNA molecules are labeled with a fluor-ophore. By using a variety of different dyes in a single multiplexed reaction, different loci can be targeted, amplified, and labeled with specific dyes. The dyes used in these reactions absorb at similar wavelengths, but emit at different wavelengths. Thus a single laser can be used to excite four or more dyes. A multichannel analyzer is then used to identify the specific PCR product by means of the wavelength of emission of the bound dye.
The development of methods for data analysis by CE is of particular importance in the examination of PCR products. Precise and reliable methods must be developed for product analysis. Slabgel methods permit the analysis of multiple samples run concurrently. CE is a serial technique; samples are run one at a time. Thus, comparison of multiple samples requires the addition of internal standards to correct for the inevitable variations in injection, temperature, and current. This observation is particularly relevant in quantitative methods where variations in sample injection can limit the usefulness of the technique.
Size estimates of PCR products can be performed by interpolation of product size based on the migration of one or more internal standards. For products in the size range 100-400 bp, a linear relationship exists between size and migration time. The size of larger products may also be estimated using nonlinear curve-fitting algorithms. Specialized instrumentation has been developed specifically for DNA analysis which utilizes internal standards that have been labeled with a different fluorescence dye than that of the product. For such systems specific algorithms have been developed to deconvolute the fluorescence signals and perform size estimates
Short Tandem Repeats
Short tandem repeats or STRs are tandemly repeated nucleotide sequences 2-6 base pairs in length. The number of repeated sequences varies between individuals and results in a high degree of length polymorphism. STRs are abundant throughout the human genome, occurring at an average rate of every 6-10 kilobases. The most common form of STR is the dinucleotide repeat. These loci are unsuitable for most forensic analyses due to the presence of PCR artifacts known as ‘stutter’ bands. This phenomenon is manifested by the appearance of extra peaks one or more repeat units away from the main product and is presumably caused by enzyme slippage during the amplification process. Tetrameric repeats tend to produce less stutter than the di- or trimeric repeats and much work has been done to validate their use in forensic casework. A core set of 13 loci has been established by the Federal Bureau of Investigation for use in the Combined DNA Index System (CODIS) (Table 1).
Sample Preparation
Purified template DNA can be amplified to yield products from a single STR locus or multiple primers can be added to a single reaction mixture to products from multiple STR loci. Cocktails are commercially available for multiplex PCR reactions that include primers for as many as 15 different STR loci. The products of these reactions are labeled with as many as three different fluorescent dyes. Both the number of dyes and coamplified STRs are certain to increase. The amplified products can be denatured in de-ionized formamide that is commercially obtained or prepared on site by using an ion-exchange resin. Care must be taken when using these resins as ionic species in the formamide can inhibit the injection process. For this reason, the conductivity of the formamide should be tested and control samples should be run prior to routine analysis. The quality of the formamide used in sample preparation can also affect column longevity.
Table 1 13 STR loci approved for use with CODIS
| STR locus | Chromosome | Repeat | Number of |
| motif | repeatsb | ||
| FGA | 4 | CTTT | 18-30 |
| VWA | 12TCTA | 11-22 | |
| D3S1358 | 3 | TCTA | 11-20 |
| D21S11 | 21 | TCTA | 25-36 |
| D8S1179 | 8 | TATC | 8-19 |
| D7S820 | 7 | GATA | 6-15 |
| D13S317 | 13 | TATC | 8-15 |
| D5S818 | 5 | AGAT | 7-16 |
| D16S539 | 16 | GATA | 5-15 |
| CSF1P0 | 5 | AGAT | 6-15 |
| TPOX | 2AATG | 6-13 | |
| THO1 | 11 | TCAT | 3-11 |
| Amelogenind | X,Y | ||
a Data obtained from STRbase, published by NIST, http:// ibm4.carb.nist.gov:8800/dna/home.htm and from the Profiler and Profiler+ users manuals, Perkin-Elmer, Foster City, CA.
b The range of repeats is approximate as new alleles are constantly being discovered.
c FGA as well as other loci in this list have complex patterns of repeats. The most common is given.
d Amelogenin is a sex-linked marker which contains a six base deletion in the X chromosome.
To yield denatured fragments ready for electro-phoresis, amplified product is added to de-ionized formamide, heated at 95°C and then snap cooled in an ice bath. Some investigators have substituted de-ionized water for formamide and have achieved results similar to those obtained using formamide, however long-term DNA stability is compromised when using water as a diluent. As mentioned earlier, filtration or dialysis can be performed on the PCR product prior to mixing with formamide to increase the amount of sample injected, however, these procedures may be unnecessary for routine analysis.
Analytical Separation
Electrokinetic injection is routinely employed to apply samples onto the capillary column. Injection time may be varied within certain ranges to affect the amount of sample applied to the column without adversely affecting resolution. Varying the injection time from 1 to 10 s has been shown to increase sample input while maintaining the resolution of the system. A particular advantage of CE is the ability to quickly reanalyze overly dilute samples by simply increasing the injection time. Samples that contain excess amplified product can be either reinjected for less time or simply diluted in a mixture of formamide and size standard and rerun.
The separation media employed for analysis are typically polydimethyl acrylamide, hydroxyethyl cellulose, linear polyacrylamide or commercially prepared polymer solutions. Most forensic laboratories have opted to purchase prepared polymer solutions for reasons of quality control and simplicity of use. Varying the polymer concentration, through the purchase of the appropriate separation media or by preparation, allows the user to fine-tune resolution.
The STRs under current study contain alleles that generally differ by a four-base repeat unit, however variants which contain deletions of one or two bases can occur in these systems. As a result it is important to design separation systems which can also resolve variant alleles. In situations where increased resolution is necessary, column length or polymer concentration can be increased, however both of these procedures can greatly increase migration times, a concern for laboratories with large numbers of samples.
Monitoring the resolution of a system allows the investigator to recognize degradation in column performance or inappropriate sample preparation. As the column ages through continued use or inadequate maintenance, sample resolution will deteriorate. Samples prepared in formamide that has not been sufficiently de-ionized will also show poor resolution. Resolution can be calculated using the standard equation:
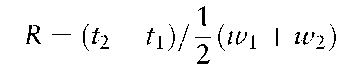
where t is the migration time of the peak and w is the peak width. Since the peak at base line is difficult to determine, the shape of the CE peak can be assumed to be gaussian with a width of 4a, and the above equation can be converted to:
![]()
where wh is the peak width at half height.
System resolution can also be quickly evaluated using a mixture containing STR alleles that vary by one base pair. The STR system THO 1 has a variant (allele 9.3) that is one base pair less than expected. This allele can be mixed in equal proportions with the nonvariant allele 10. After this mixture is run on the capillary, the evaluation of the relative height of the peaks versus the valley point between them yields a ratio that can be monitored to obtain the relative resolution of the system. Most forensic systems under current study do not provide baseline resolution of STRs that differ by one base pair, and hence monitoring the relative valley value can provide a handle on the relative performance of the system.
Genotyping
Multiple STR loci can be determined during a single analysis by adjusting the fragment length of each PCR product and by labeling sets of different primers with dyes that fluoresce at differing wavelengths. Primers are carefully designed to produce DNA with allele sizes that do not overlap. This permits the analysis of 3-4 STR loci which are labeled with the same color. Figure 3 illustrates this procedure by showing an allelic ladder consisting of all possible alleles from three different STR loci amplified together and labeled with a blue dye. Then, by using mixed sets of colored primers, 10 or more loci may be analyzed simultaneously (Fig. 4). This task is performed using CE instruments equipped with detector arrays capable of analyzing all dyes simultaneously.
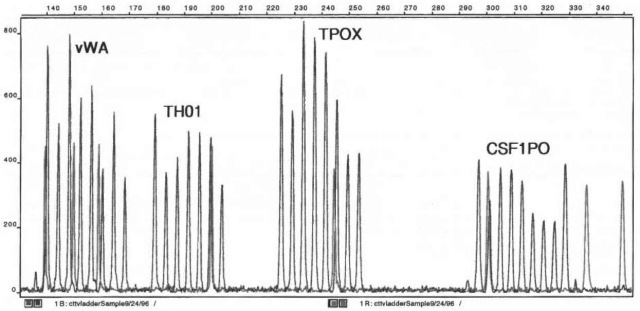
Figure 3 An allelic ladder from the Geneprint cttv multiplex (Promega) consisting of alleles from three genetic loci, CSF1P0, TPOX, TH01, and vWA. The internal standard (labeled in red) is a Genescan 350 ROX (Perkin-Elmer) size standard. Analyzed using an PE/ABI 310 capillary electrophoresis system with laser induced fluorescence detection and a hydroyxethyl cellulose sieving matrix.
The peaks resulting from this analysis can be genotyped through the use of software supplied by the instrument manufacturer. Typically, electrophor-esis is conducted by combining amplified products with an internal size standard that is derivatized with a fluorescent dye that is different from those used to label the STR loci. Figure 3 illustrates this technique. In this figure the red size standard provides an internal reference to standardize the electrophoretic run and permits the calculation of the base pair sizes of the detected peaks. The calculated sizes can be compared to those sizes obtained from the allelic ladders, which are run separately. The allelic ladder is used to calibrate a series of electrophoretic runs, much like the internal standard used to standardize a single run.
The resulting data are compared to tables containing the frequency of each allele in a target population. The frequency of a particular profile can then be calculated by multiplying together the component frequencies calculated for each locus. The resultant frequencies can be quite small. For example, one commercial set of 10 loci has a probability of identity of 1 x 10″11.
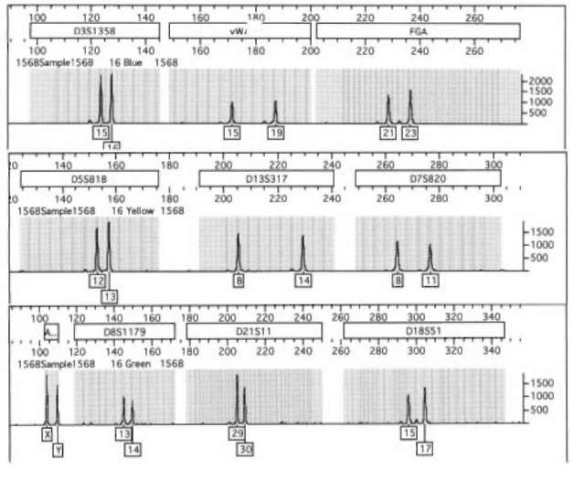
Figure 4 A single analysis of a blood sample amplified using the Profiler+ (Perkin-Elemer) PCR amplification kit. The results have been split into three panels to aid in analysis. Each gray panel indicates a different STR locus and is identified above the panel. The dark gray zones within the lighter gray indicate potential locations of alleles for each locus. Note the small stutter peaks to the left of certain of the larger peaks. Conditions: PE/ ABI 310 genetic analyzer using POP4 sieving matrix and laser induced fluorescence detection.
Mixture Analysis
Mixtures, samples that contain DNA from more than one individual, must be anticipated in the analysis of forensic specimens. These specimens may be a composite of body fluids from different individuals and will produce complex DNA profiles. To complicate the analysis of mixtures, STR patterns from one individual may contain imbalanced peaks and PCR artifacts known as stutter.
Stutter peaks are amplified products resulting from the ‘slippage’ of DNA polymerase during amplification where the enzyme and growing DNA chain are out of alignment with the target DNA. The resulting fragment is usually 4 bp less than the true allelic peak, although weaker signals consisting of sets of peaks four bases apart may also be seen. Some loci have yielded stutter peaks of > 10% of the height of the true peak. In the interpretation of mixtures, the possibility of stutter peaks must be taken into account and interpretations should be adjusted based on the amount of stutter observed at a particular locus. Typically, a mixture is suspected when peak heights rise above typical stutter values for a particular locus.
Another problem in the interpretation of mixtures is that peaks obtained from a heterozygous locus may vary by as much as 30%. Deviations of this size, although uncommon, must be considered in the evaluation of mixtures. Differences in the expected peak ratio in a sample that presents a heterozygous pattern, can indicate a mixed sample whose alleles have co-electrophoresed. Fortunately, when multiple allelic systems are evaluated, other loci may show three or four peaks, establishing the specimen as a mixture, and can be used to determine if the altered ratio could be due to an overlapping allele. Figure 5 illustrates the analysis of a sample of mixed DNA.
The balance observed between loci is an additional consideration in the assessment of mixtures. Although commercially available STR amplification kits attempt to achieve a balance, as the size of the template required for amplification increases, the amount of amplified product typically decreases. In situations where degraded samples are used, shorter PCR products will predominate as few of the longer fragments of template have survived. Under such circumstances, peaks that represent a minor component in a short locus may not be present in other larger-sized loci. Nonallelic peaks occasionally also cause problems in the interpretation of a mixed sample. These peaks may be the result of free dye, unbound dye, electrical interferences or other sample artifacts. There are also PCR artifacts which occur resulting in a peak one base pair less than the true allelic peak. In most STR systems the amplification process promotes the nontemplate addition of a nucleotide (usually an A) to the end of the PCR product. This yields an amplicon that has been increased in size by one base pair. Under some conditions, such as excessive amount of template DNA or Taq inhibitors, the complete conversion of all the products to the «+1 state may not occur. Often this condition can be rectified by increasing the extension time for the amplification.
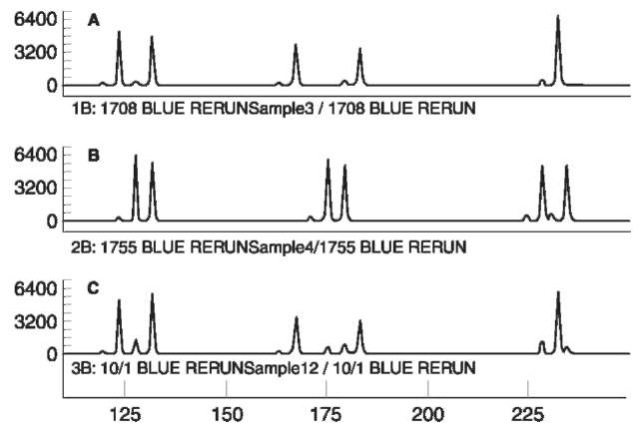
Figure 5 The analysis of a mixed sample of DNA using the AmpFISTR blue STR kit (Perkin-Elemer). Panels A and B contain individual samples. Sample C is a mixture with sample B as the minor component. Note the effect of stutter peaks on the relative size of the alleles. Conditions as in Figure 4.
Finally, mutations or rare genetic events, may give rise to unusual profiles that can be mistaken as a mixture. In addition, mutations at primer sites may lead to genotyping variations at particular loci, due to differences in the location of primer annealing sites used by the various manufacturers of STR amplification kits.
Databasing
For many years most USA DNA laboratories did not pursue the analysis of cases suitable for DNA analysis unless standards for comparison were obtained. With the introduction of a national DNA database, this policy has changed. Now many laboratories are preparing to profile specimens in cases where no suspect exists in the hope of identifying the suspect through a database search. The FBI has established CODIS as a database to allow for the exchange and comparison of DNA profiles. All 50 states have legislation that enables them to collect DNA specimens from individuals convicted of certain crimes. When profiled, CODIS will act as the repository for the convicted offender profiles so that laboratories linked to the database can access the files. In addition to the database of convicted offenders, CODIS will also permit the comparison of profiles generated from crime scene samples that would allow investigators to link crimes to repeat offenders. Many crimes have been solved or linked through database searches, which in the USA have been primarily RFLP (restriction fragment length polymorphism) based. With the advent of STRs, and given the relative ease of analysis compared to RFLP, more samples of both convicted offenders and crime scenes will be profiled and entered into the database.
Analysis of Mitochondrial DNA
There are circumstances in forensic analysis in which there is insufficient nuclear DNA to perform PCR. These cases involve samples such as shed hairs or those that are highly degraded. In these circumstances there may still be enough mitochondrial DNA to permit PCR amplification. The DNA present in mitochondria is approximately 16 000 bases long and contains a section known as the control region that contains a number of polymorphic sites which are usually point mutations. In this procedure, certain hypervariable segments of the control region are PCR amplified and sequenced. These sequences are then compared to a known standard in order to identify polymorphic sites.
CE is used in this process to determine if the amplified product is present in sufficient quantity and purity for the sequencing step to be practical. A small portion of the amplified product is analyzed in its native state using a short 27 cm capillary at 15 000 V. In the analysis, an intercalating dye is added to the detector to provide a fluorescent product and a result is obtained in under 4 min. The peak intensity of the amplified DNA is compared to an internal standard to determine the quantity of amplified material. Figure 6 illustrates this separation. The electropherogram is also checked to determine if any contaminants such as primers or extraneous amplified product is present in the sample. These materials can interfere with the sequencing reactions and reduce the quality of the result. The results of this analysis are used to adjust the amount of template used for the sequencing reaction. The products are then analyzed on gel-based sequencers. However, as sequencers based on CE become more widespread, both the product analysis and the sequence analysis will be performed via CE.
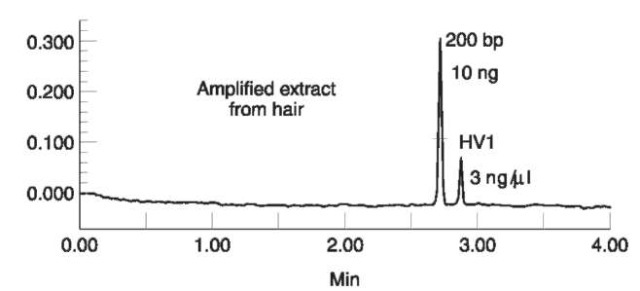
Figure 6 The analysis and quantitation of mitochondrial DNA in under four minutes. The quantity of DNA produced is determined with reference to the internal standard. Conditions: Beck-man PACE capillary system with laser induced fluorescence. 27 cm DB-17 capillary run at 15 kV and filled with 1% hydroxy-ethyl cellulose and 50ng/ml YO-PRO-1 in TBE buffer. Figure courtesy of Dr. John Butler, GeneTrace Systems.
There are also a number of other methods for detection of point mutations in mitochondrial DNA for which CE can be used. These include single-strand conformation polymorphism (SSCP) and PCR-RFLP. These techniques can provide a quick comparison of known and questioned samples of the amplified products. If the patterns produced do not match, then an exclusion has occurred and it is not necessary to perform the sequencing step.
Future Applications
Unlike slabgel-based systems, CE instruments must inject samples in a sequential fashion. However, it is possible to perform the capillary experiment using a bundle or array of individual capillaries to inject multiple numbers of samples at the same time. Such systems can inject 96 samples or more at the same time. Then, using software algorithms, the migration times from each individual capillary are scanned, synchronized using internal lane standards, and the data are reported. Capillary array electrophoretic systems can produce tremendous amounts of data in a relatively short time.
Recognizing the increased complexity required by the capillary array systems, a number of researchers are developing smaller, more compact systems based on microchip technology. By using photolithography, multiple channels can be etched into silicon wafers and the entire capillary array can be placed on a silicon chip. Tightly focused sample injections on a microchip permit fast electrophoretic separations using relatively short channels. A further advantage of this technique is that sample preparation and detection apparatus can be built into the chip design. Thus the entire process from DNA extraction to PCR to separation to detection can be integrated into a single device.
Conclusions
Capillary electrophoresis is a technique which provides the DNA analyst with much flexibility. CE systems utilize replaceable physical gels which are pumped into the capillary at the beginning of each analysis. DNA quantitation, genotyping and sequencing are all possible using this technique. Sample injection, separation and analysis can be easily automated. Multichannel fluorescence detection permits multiplex PCR reactions to be simultaneously analyzed, greatly conserving precious forensic samples. The resulting data can be analyzed to detect the presence of mixtures, processed, and stored in a database known as CODIS. Present systems under development will utilize arrays of capillaries to increase sample throughput. Future integration of the entire process of DNA analysis is possible using CE due to the distinct advantages of this technique.
