D. Triosephosphate Isomerase
Triosephosphate isomerase (TIM) catalyzes the inter-conversion of D-glyceraldehyde-3-phosphate (G3P) and dihydroxyacetone phosphate (DHAP). The equilibrium lies far to the side of DHAP, hence the longer arrow pointing to that compound. The enzyme operates with a turnover number of ~107 s-1, which is nearly as fast as the diffusion-controlled limit. TIM is therefore called an almost perfectly evolved enzyme because no catalytic refinement could make the rate faster than it already is.
Many tools have been used to study the TIM mechanism, including X-ray crystallography and NMR, site-directed mutagenesis, and affinity labeling. Strong evidence for the mechanism, however, was supplied by studies using isotopic labeling of substrates. It was found that if the above reaction was carried out in tritiated water, one atom of tritium was stereospecifically incorporated into DHAP.
This result suggested that a base abstracts a proton from the substrate, and the proton then undergoes exchange with labeled protons from the solvent before being added back to form the product stereospecifically. The existence of a cis-enediol intermediate (shown below) would account for these observations, if the enzyme added the proton back to the same face of the enediol that it was abstracted from.
One unresolved question was whether only a single protein base was involved (so that the transfer was from substrate to base and directly back to form product) or whether a different base was responsible for protonation as part of a more extensive proton relay. The nature of the protein base was explored by doing a similar experiment to the one described above but in the other direction; that is, by labeling the DHAP and observing its conversion to G3P. Although the equilibrium lies far to the side of the DHAP, trapping by irreversible oxidation by G3P dehydrogenase of any G3P formed was used to convert significant quantities of DHAP. If the DHAP was labeled at C1, a small but measurable amount of the label was transferred to C2.
This result suggested that a single base was involved in the proton abstraction/proton addition step. If more than one base were involved, the chance that any label would not be washed out by the solvent and would be added to the deprotonated intermediate would be vanishingly small. In combination with other kinds of experiments, isotopic labeling was therefore invaluable in elucidating the mechanism of triosephosphate isomerase (shown below) in which B is a protein-derived base.
Isotopes can be used in another way to measure the energy barrier heights for various steps in the catalytic mechanism as noted above for the reaction catalyzed by dihydrofolate reductase. For example, if a proton transfer is involved in the rate-limiting step, then substitution of that proton with one of the heavier isotopes of hydrogen (deuterium or tritium) will cause the step to proceed more slowly. These so-called kinetic isotope effect experiments in combination with steady-state rate measurements in the case of TIM allowed the elucidation of the rate constants for partitioning of the cis-enediol intermediate and construction of a detailed kinetic scheme as shown above for dihydrofolate reductase.
E. Aspartate Aminotransferase
Many enzymes employ exogenous molecules known as cofactors to assist in executing their chemistry. Sometimes these cofactors are covalently bound to the enzyme and sometimes not. Many types of cofactors are known, and here we will focus on a well-studied example called pyri-doxal phosphate (PLP), which often participates in the metabolism of amino acids. PLP, derived from vitamin B6, is a covalently bound cofactor; it is attached to lysine residues by means of a Schiff base or imine linkage as shown at right.
The substrates for most PLP-requiring processes are a-amino acids, and most of the processes take place at the a-carbon position, although some take place at the j- or y-carbon. The enzymes which use PLP catalyze a wide range of reactions, including racemizations, decar-boxylations, and amine transfers. In general, for all three of these classes of reactions at the a-carbon the substrate displaces the lysine and forms an aldimine intermediate with the PLP.
The now very acidic a-proton of the amino acid is abstracted by a basic amino acid residue (often the displaced lysine), with the pyridine ring of PLP acting as an electron sink. For the racemases, a proton is then delivered to the opposite face from the same or a different basic residue with the net result of inversion of configuration at the a-carbon. Attack of the active site lysine effects product release and regenerates the cofactor.
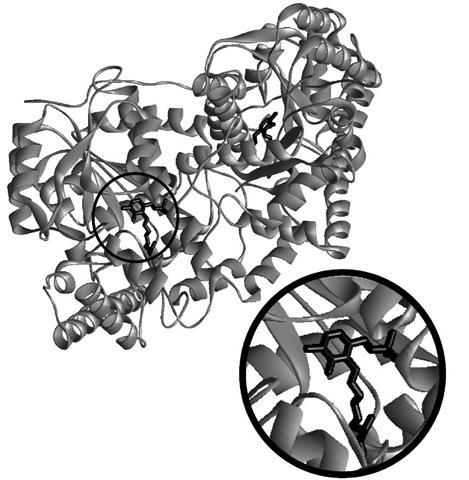
The structure of one PLP-utilizing transaminase, aspartate aminotransferase, is shown in Fig. 8. This enzyme catalyzes the reversible transamination reaction shown below.
FIGURE 8 Structure of an aspartate aminotransferase. The protein is a homodimer, with one covalently bound pyridoxal phosphate (shown in black) in each of the two subunits. The expanded view shows the cofactor in greater detail.
In the transamination reaction, formation of the aldi-mine intermediate between aspartate and PLP and its deprotonation proceeds as described above for the race-mases. However, reprotonation occurs not at the same carbon as in the racemization mechanism but at a position adjacent to the PLP heterocycle.
Hydrolysis releases the product oxaloacetate and generates a new form of the cofactor called pyridoxamine. The reverse reaction is then carried out on the other substrate, a-ketoglutarate, forming glutamate and regenerating the PLP cofactor.
Reactions at the j -position (for example, in threonine dehydatase) or the y-position (in methionine-Y-lyase) also proceed by means of formation of an aldimine intermediate with the a-carbon of an a-amino acid. Such a survey of PLP-dependent enzymes illustrates the important point that one cofactor can be used for different kinds of transformations. The reactions described all go through a common aldimine intermediate, with the ultimate course of the reaction being controlled by the appropriate substrate specificity and positioning of amino acid side chains. This flexibility allows nature to expand its chemical repertoire with a relatively small set of cofactors.
There are other organic cofactors such as thiamine pyrophosphate and biotin that participate in carbon-carbon bond formation and cleavage, cofactors that participate in reduction/oxidation, or redox, reactions such as nicotinamide and flavin moieties discussed in some of the earlier examples, and still others that are metal based such as vitamin B12 and porphyrin, which is our next topic.
F. Cytochrome P450
A different kind of cofactor from PLP is responsible for the chemistry of cytochrome P450 (Fig. 9), an enzyme which oxidizes hydrocarbons. It is known as a mixed-function oxidase, or monooxygenase, because one oxygen atom from molecular oxygen is incorporated into the product while the other goes on to form water. Cytochrome P450 in the liver, for example, oxidizes and detoxifies many kinds of substances that would otherwise be poisonous. One such well-studied reaction, the hydroxylation of camphor, is depicted below.
In the structure in Fig. 9, the iron porphyrin is shown in black.
Cytochrome P450 is a redox catalyst. The multiple available oxidation states allow the cofactor to accept and donate electrons during different stages of the catalytic cycle. Since the early 1970s, this enzyme and its relatives have been the subject of intense study by, among others, enzymologists, toxicologists, biophysical chemists, and inorganic chemists. The latter have tried to model the chemistry of cytochrome P450 with synthetic small molecules with the twin goals of mimicking its activity and understanding how the enzyme itself works. Working in parallel, biophysical chemists and enzymologists have performed many steady-state and pretransient kinetic studies such as the ones already discussed, which have contributed to a working model for the mechanism shown in Fig. 10.
Although this mechanism is in some senses more complicated than those that we have discussed, the same concepts apply. Starting at the top of the cycle, in the resting state of the enzyme the iron is in the +3 oxidation state and is bound by water. Substrate docks to its specific binding site and displaces water to start the catalytic cycle, and an electron is then introduced to reduce the iron to the +2 oxidation state. The dashed line is meant to indicate association of the substrate with the active site, not an actual bond to the iron. The requirement that substrate bind before reduction occurs is a control feature which prevents formation of very active and potentially damaging species in the absence of substrate. Oxygen then binds and accepts an electron from the iron, and introduction of another electron and two protons allows one atom of dioxygen to be released as water, which leaves behind a very active high valent (formally iron 5+) species. What follows is known as a radical rebound step. A hydrogen atom is removed from the substrate and transferred to the terminal oxygen atom, which produces a substrate radical. The radical recombines with the new hydroxo moiety to form the hydroxylated product, which is then displaced by water; this completes the catalytic cycle.
At the core of cytochrome P450 is an iron porphyrin, or heme, group, protoporphyrin IX, which is depicted below.
One important line of investigation which has supported the radical rebound hypothesis is the use of radical clock substrate probes. These probes rearrange in a diagnostic way on a very rapid and calibrated time scale when a hydrocarbon radical is formed. In the case of P450, rearranged products have been isolated after oxidation and have been used as evidence of an intermediate substrate radical. In this way, even though the lifetime of the radical is too short for it to be observed directly, its character can be explored by the judicious choice of substrate analogues.
Mechanistic proposals are under constant scrutiny and revision, and aspects of the foregoing mechanism have been challenged. In particular, the possibility has been suggested that a species other than a high valent iron-oxo (likely a hydroperoxo species) may be the active oxidant for some substrates. Debates such as these are a great strength of the study of enzyme mechanisms. Given all the tools which have been developed in this field, and the wealth of interesting problems to which these tools can be applied, the study of enzyme mechanisms should be considered a vital and evolving process. The answer to the question of "how enzymes work" cannot be described fully in a single scheme.
![tmp4647_thumb[2] tmp4647_thumb[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp4647_thumb2_thumb.jpg)
![tmp4648_thumb[2] tmp4648_thumb[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp4648_thumb2_thumb.jpg)
![tmp4649_thumb[2] tmp4649_thumb[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp4649_thumb2_thumb.jpg)
![tmp4650_thumb1[2] tmp4650_thumb1[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp4650_thumb12_thumb.jpg)
![tmp4651_thumb[2] tmp4651_thumb[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp4651_thumb2_thumb.jpg)
![tmp4652_thumb[2] tmp4652_thumb[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp4652_thumb2_thumb.jpg)
![tmp4653_thumb1[2] tmp4653_thumb1[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp4653_thumb12_thumb.jpg)
![tmp4654_thumb1[2] tmp4654_thumb1[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp4654_thumb12_thumb.jpg)
![tmp4656_thumb1[2] tmp4656_thumb1[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp4656_thumb12_thumb.jpg)
![tmp4657_thumb[2] tmp4657_thumb[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp4657_thumb2_thumb.jpg)
![tmp4658_thumb[2] tmp4658_thumb[2]](http://what-when-how.com/wp-content/uploads/2011/05/tmp4658_thumb2_thumb.jpg)
