1. Introduction
Worldwide research efforts to characterize the human genome, such as the Human Genome Project (see Article 24, The Human Genome Project, Volume 3), the ENCODE project, and the International HapMap Project, along with advances in DNA sequencing technologies, have produced an enormous amount of DNA sequence information. This information is becoming available in public databases and will allow researchers to identify and characterize naturally occurring variations in the human DNA sequence across individuals. Such genetic variation, that is, differences in DNA sequence among a group of individuals or between populations, occurring with frequency >1% is known as genetic polymorphism. Sources of polymorphisms within DNA sequence include variable numbers of tandem repeats (VNTRs) such as microsatellite repeats and short tandem repeats (STRs), small insertions and deletions of sequence (in/dels), gene copy number variation (Sebat et al., 2004; Fredman et al., 2004), and single-nucleotide polymorphisms (SNPs) (Kwok et al., 1994). SNPs are the most abundant form of genetic polymorphisms found in the human population and have become a focal point in the study of the genetic basis of multifactorial diseases and traits (see Article 57, Genetics of complex diseases: lessons from type 2 diabetes, Volume 2), interindividual differences in response to therapeutic drugs (“pharmacogenomics”), and human evolutionary and population history (see Article 71, SNPs and human history, Volume 4). This article will outline sequencing-based molecular and bioinformatics approaches for the identification of SNPs in the human population. Although approaches discussed can be extended to study polymorphism in other organisms and/or other types of polymorphism, the sequencing-based identification and scoring of repeats and in/dels can be more challenging than for SNPs.
As noted, SNPs are the most simple and abundant form of a genetic polymorphism and occur along a stretch of DNA sequence when one of the four nucleotides that contain the base adenine(A), cytosine(C), thymine(T), or guanine(G) is replaced by one of the remaining three nucleotides. A SNP that occurs between the purines (A, G) or pyrimidines (C,T) is known as a transition and constitutes two-thirds of all SNPs (Iida etal., 2001a,b,c; Freudenberg-Hua etal., 2003); a transversion occurs between a purine and a pyrimidine.
By 2001, data arising from the public Human Genome Project identified over one million SNPs averaging one SNP every 1331 bp in comparison of two human chromosomes drawn from an ethnically diverse sample of individuals (The International SNP Map Working Group, 2001). A combination of polymorphisms that are closely linked on a single chromosome and that are inherited together on a single maternal or paternal chromosome is termed a haplotype. SNPs have become instrumental in defining haplotype “blocks” that reflect stretches of genome encompassing multiple SNPs with alleles that appear to be inherited together in the human genome (The International HapMap Consortium, 2003; http://www.hapmap.org).
2. In silico SNP discovery
Numerous SNPs were initially detected by comparing sequences from independent sources of mRNA and expressed sequence tags (ESTs) that had either been deposited in the public domain (dbEST; Boguski etal., 1993) or generated by large sequencing projects (Marth et al., 1999; Irizarry et al., 2000; Useche et al., 2001). Over 48 000 SNPs were identified through the use of purely computational and bioinformatic tools. This “in silico” mining of SNPs consist of simple comparisons of extant DNA sequence obtained from different sources, individuals, or investigators. These SNPs have been integrated into public databases such as dbSNP at the National Center for Biotechnology Information (NCBI). Owing to the potential errors in sequence information, either introduced by Taq polymerase during PCR or computational errors associated with base calling algorithms necessary for interpreting the results of sequence assays, in silico-defined SNPs should be validated through direct sequencing to confirm their existence in the human population. NCBI’s dbSNP catalogs over 900 000 SNPs (Sherry etal., 2001) as well as small in/dels, and retroposable element insertions; other SNP databases contain SNPs specific to particular ethnic populations (e.g., JSNP), while others catalog SNPs involved in diseases (e.g., HGVSNP) (Table 1). SNP databases are a good resource for investigators to verify SNPs that they have identified in their own sequencing studies, for choosing loci for further sequencing studies, and for selecting SNPs to genotype for association studies. However, the investigator should be aware that SNPs contained in these databases have been obtained through various techniques, including in silico approaches, and a number of these SNPs remain unvalidated while others may have been missed because of the technique used or the population studied. Recent studies have investigated the quality of SNP databases (Jiang et al., 2003; Reich et al., 2003; Mitchell et al., 2004).
3. Targeted polymorphism discovery at candidate loci
Computational mining of SNPs from available DNA sequence databases has led to the identification of only a fraction of the SNPs’ existing in the population; rare SNPs having low allele frequencies, or SNPs that are unique to a specific population (“population specific SNPs”) are less likely to be found by these approaches (Kruglyak and Nickerson, 2001). Direct sequencing of a population of individuals followed by a comparison of the resulting sequences through multiple sequence alignment procedures is the most reliable of methods for identifying SNPs and other polymorphisms.
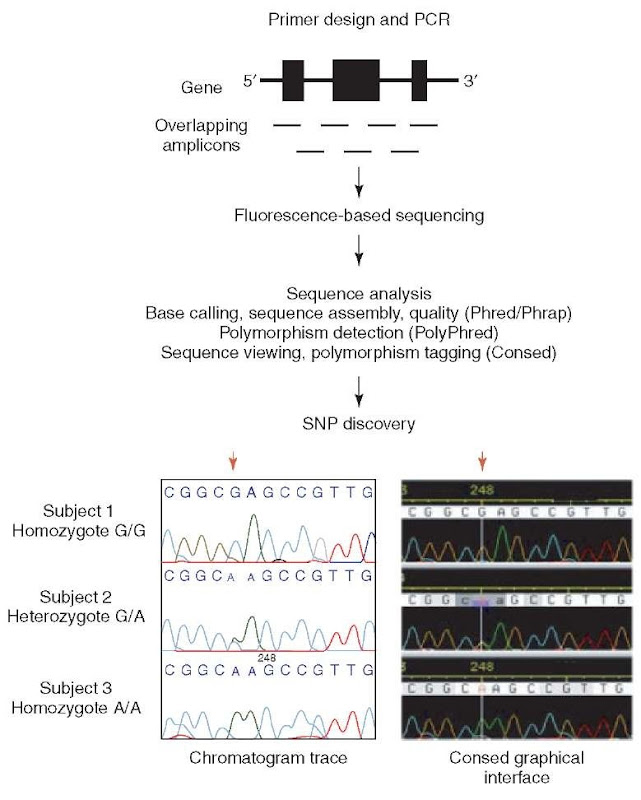
Fluorescence-based sequencing has become an important tool for such polymorphism discovery studies (see Griffiths et al., 1996, for a description of available sequencing methods). Figure 1 summarizes the steps in sequence-based polymorphism discovery following the selection of samples for sequencing: Polymerase chain reaction (PCR) amplification of locus in samples; sequencing of PCR products from forward and reverse directions; base calling, contig assembly, and polymorphism detection by sequence analysis software; and finally visualizing aligned sequence chromatograms for polymorphism identification and heterozygous base calling.
3.1. Samples
The choice of DNA samples depends on the ethnic population of interest, trait, or disease being studied, and the power desired to detect a polymorphism of a certain frequency. For example, sequencing 50 individuals (100 chromosomes) will enable the identification of SNPs that are at least 1% in that population with high (>80%) confidence. The frequencies of a number of SNPs are known to vary in different populations. Thus, limiting studies to a single population will reduce the chance of identifying SNPs common in other populations that are frequent or rare in the studied population. Therefore, to identify disease-associated variants, sequencing DNA from a pool of affected individuals is ideal, and ethnic background should be considered (see Article 75, Avoiding stratification in association studies, Volume 4).
However, to maximize the probability of finding a variant in the general human population, ethnic diversity panels is the best strategy. To facilitate the discovery of genetic variants in the entire human population, the National Human Genome Research Institute (NHGRI) of NIH, in conjunction with the Centers for Disease Control and Prevention, the National Institute of Environmental Health Sciences, and individual investigators, has assembled a DNA Polymorphism Discovery Resource consisting of DNA samples from 450 unrelated individuals from the United States, with ancestry from major regions of the world (Collins et al., 1998).
Figure 1 Targeted polymorphism discovery at candidate loci. PCR primers are designed to generate overlapping fragments (amplicons) of DNA to accommodate sequencing the entire gene of interest. Fluorescence-based sequencing generates chromatograms that are analyzed through the software package of choice to facilitate polymorphism discovery. Consed tags an SNP at position 248. A red arrow above both chromatogram views indicates the location of the discovered SNP. Subjects 1 and 3 are homozygous for nucleotide G and A, respectively. Subject 2 is determined to be heterozygote based on overlapping G and A traces that have approximately one-half the amplitude of either homozygote peak
These samples, as well as the NIH Diversity Panel, are available from the Coriell Institute for Medical Research (http://coriell.umdnj.edu) in collaboration with the National Institute of General Medical Sciences (http://locus.umdnj.edu/nigms/).
3.2. PCR amplification of DNA samples and sequencing
Forward and reverse sets of oligonucleotide primers for PCR amplification of a desired region of the genome are designed on the basis of gDNA sequence obtained, for example, from GenBank (NCBI). Messenger RNA sequence does not contain intronic sequence found in gDNA, so care should be taken when designing PCR primers on the basis of these edited sequences for gDNA templates. Depending on the sequencing assay and instrument used, PCR primers should be designed to amplify approximately 600-1000-bp fragments.Overlapping at least 100 bp on each end of the PCR fragment will reduce the chance of missing SNPs located in the primer region or on the ends of the PCR-amplified fragments (amplicons), which generally do not sequence well by fluorescence-based methods. Following the PCR amplification reaction, amplicons are purified and prepared for sequencing. The set of forward and reverse PCR primers for each amplicon can be used as sequencing primers to sequence from both ends of the amplicon. Alternatively, sequencing primers can be designed on the basis of other sequences contained in the amplicon.
3.3. Sequence analysis
Accurate and efficient sequence analysis for fluorescence-based sequencing requires software for base calling, sequence assembly, polymorphism detection, and sequence visualization. Two examples of reliable software packages for sequence analysis are MacVector (Rastogi, 2000) and the Phred/Phrap/Consed suite of sequence analysis software, the latter of which we present here in more detail. Phred is used to automate base calling including accuracy assessments of base calls (Ewing et al., 1998; Ewing and Green, 1998); Phrap assembles sequence fragments of multiple amplicons on the basis of a representative sequence of the locus; and Consed enables the visualization of sequence alignments as well as the original fluorescent graphical data (“chromatogram traces”) from which the base calls were made (Gordon et al., 1998). Polymorphisms called by sequence analysis software should also be reverified manually using a graphical interface such as Consed.
3.4. Heterozygous base calling
Since humans are diploid (have two copies of each chromosome – one inherited from the mother and one inherited from the father), a DNA sequence from gDNA template represents both copies of the chromosome. An individual is heterozygous at a polymorphic site when two different alleles (two different bases for a SNP) exist at that site. It can be difficult to accurately identify heterozygous sites because of the variability in fluorescence signals and inconsistency of base calling at these sites. The detection of heterozygous sites when comparing sequence traces of homozygotes with heterozygotes is based on the observation of a significant drop in peak height (to approximately one-half) along with the presence of a second, overlapping peak in heterozygotes. PolyPhred is one of the most reliable software packages available for automated detection of heterozygous sites (Nickerson et al., 1997). Chromatogram traces from multiple individuals may be compared, for example, with Consed, to help assess difficult heterozygous calls.
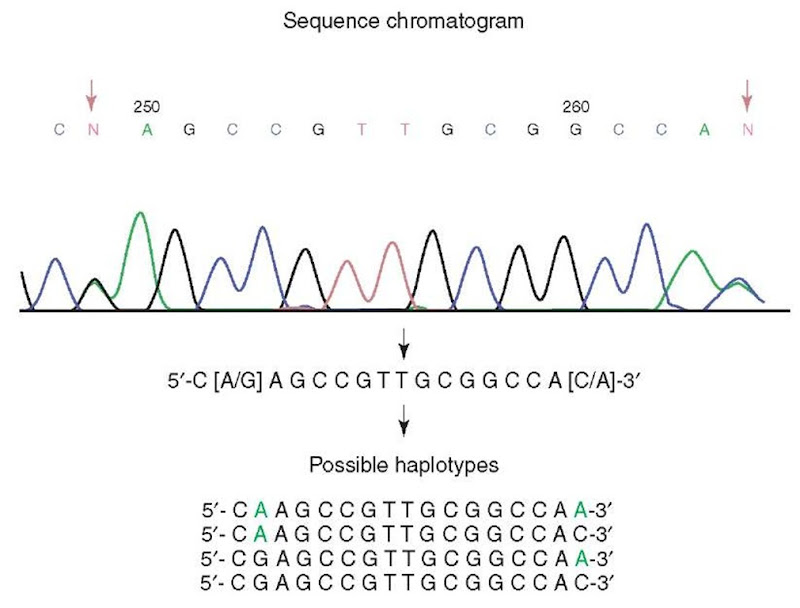
Figure 2 Complexity of haplotype determination in diploid organisms. A segment of continuous sequence from a human subject has two SNP sites, position 249 G/A and position 264 A/C. There are four possible combinations of two SNPs representing four possible haplotypes. All four haplotypes or only a subset may be represented in the human population. Further analysis is needed to determine the true haplotypes of this subject
3.5. Verifying rare genetic variants
The advantage of polymorphism discovery studies on a large number of samples is being able to identify rare variants in the population. However, genetic variants observed only once should be verified through reamplification from original gDNA template followed by sequencing to rule out the possibility of Taq polymerase induced or other sequence analysis errors.
3.6. SNP haplotype assembly
Because of the predominant diploid character of the human genome (see above), SNP identification in fragmented sequences introduces an algorithmic problem for assembly of multiple SNP genotypes into true haplotypes. If there are two or more SNPs in a fragment of genomic sequence, it is impossible to determine the haplotype just from that sequence.The most reliable method for delineating haplotypes is to clone a DNA fragment containing multiple SNP sites into a bacterial plasmid vector. After transformation of cloned vector into bacteria, a single colony representing the sequence from one chromosome can be chosen for sequencing. (see Griffiths et al., 1996, pp. 424-432). The sequence of the fragment representing the other chromosome can be accurately inferred on the basis of the sequenced chromosome fragment. When multiple individuals are studied, haplotyping each individual using the cloning method is time consuming and costly. In such cases, algorithms that statistically infer haplotypes may be a more efficient approach (Table 2). When the number of individuals is large or family data exists, such statistical inference can be reasonably accurate (Salem et al., 2005).
4. Conclusion
Polymorphism discovery relies heavily on sequencing-based methods of detection as presented here, however, other nonsequencing-based alternatives have been used. For example, the denaturing high-performance liquid chromatography (dHPLC) method, which utilizes the different mobility of homo- versus heteroduplexes of PCR amplicons during liquid chromatography under partial denaturing conditions, has successfully been employed for SNP discovery (Han et al., 2004). Such nonsequence-based genotype detection methods must be followed up with sequencing to determine the actual sequence change of the variant. Once SNPs and other polymorphisms have been discovered, one of the alternate high- or moderate-throughput strategies for genotyping large numbers of individuals can be employed as these will be more efficient than direct sequencing when the investigator is interested in typing subjects at single polymorphic sites (reviewed in Kwok, 2001; Gut, 2001).