1. Introduction
Gene prediction is the first and most fundamental step to genome analysis and annotation (see Article 13, Prokaryotic gene identification in silico, Volume 7, Article 14, Eukaryotic gene finding, Volume 7, Article 21, Gene structure prediction in plant genomes, Volume 7, and Article 26, Dynamic programming for gene finders, Volume 7). Consequently, the development of gene-finding software continues to be an important area of research in Bioinformatics. Gene-structure prediction is particularly challenging for eukaryotes, in which the density of genes is low and most genes consist of multiple exons separated by introns of varying length; see Guigo (2004) for an overview on existing software for gene prediction in eukaryotes. During the last years, a large number of computer programs have been developed to identify genes in eukaryotic genome sequences. Recent studies show, however, that the reliability of these methods is limited. While most methods work well on small sequences with single genes, their performance decreases considerably when applied to BAC-sized sequences including multiple genes (Guigo et al., 2000).
Traditional gene-prediction programs rely on information derived from known genes. Intrinsic methods use statistical features such as ORF length or codon usage and try to identify splice junctions and other signals. The underlying models are trained using previously known genes from the same or a closely related organism. Traditional extrinsic methods work by comparing genomic sequences to EST or protein sequences.
With an ever-increasing number of completely or partially sequenced genomes, new comparative approaches have been proposed to predict genes and other functional elements. The idea is that functionally important parts of the genome are under selective pressure, so they are usually more conserved than nonfunctional regions where random mutations can be tolerated without affecting the fitness of the organism. Consequently, local sequence conservation among genomic sequences usually indicates biological functionality. This phylogenetic footprinting principle has been successfully applied to various problems such as detection of regulatory elements, and it is now generally accepted that comparative sequence analysis is a powerful and universally applicable tool for functional genomics.
Gene prediction by comparative sequence analysis means that instead of comparing a given sequence to previously known genes, one compares genomic sequences from evolutionarily related organisms to each other. Regions of local sequence conservation are then searched for relevant signals to identify possible exons and gene structures. Note that, in contrast to traditional approaches, comparative gene finding is possible without any previous knowledge about existing genes and their statistical composition – except for simple models of splice signals. Therefore, these new approaches are able to detect genes with unusual statistical features that can be missed by traditional methods.
2. Gene prediction using CHAOS, DIALIGN, and AGenDA
Comparative gene-finding approaches have been developed, for example, by Bafna and Huson (2000), Batzoglou et al. (2000), and Wiehe et al. (2001). At MIPS/GSF research center and at the University of Bielefeld, we developed a comparative gene finder called AGenDA (Alignment-based Gene-Detection Algorithm, Rinner and Morgenstern, 2002). The first step in comparative sequence analysis is to calculate an alignment of the sequences under study as the results of any comparative method can be only as good as the underlying alignment. For AGenDA, we used the alignment program DIALIGN (Morgenstern et al., 1996). This program integrates local and global alignment features by constructing alignments based on pairwise local homologies. In a number of recent research projects, DIALIGN has been applied to identify functional elements in genomic sequences.
In a preliminary study, we found that local homologies identified by DIALIGN in genomic sequences are highly correlated to protein-coding exons (Morgenstern etal., 2002); in this regard, DIALIGN proved to be superior to alternative alignment programs. However, since DIALIGN has originally been designed as a general-purpose aligner, the standard version of the program is too slow to align large genomic sequences. Thus, we implemented an anchored-alignment approach to speed up the alignment procedure. Here, we are using the fast database search tool CHAOS to identify local sequence similarities; these similarities are then used as prealigned anchor points to reduce the alignment search space and program running time for DIALIGN, as described by Brudno et al. (2003).
On a given pair of input sequences, for example, from human and mouse, AGenDA performs the following operations (for details, see Rinner and Morgen-stern, 2002):
1. High-scoring sequence similarities identified by CHAOS and DIALIGN are clustered by bridging small gaps between them.
2. Conserved splice junctions and start/stop codons are identified around the cluster boundaries. For splice signals, standard matrices proposed by Salzberg (1997) are used.
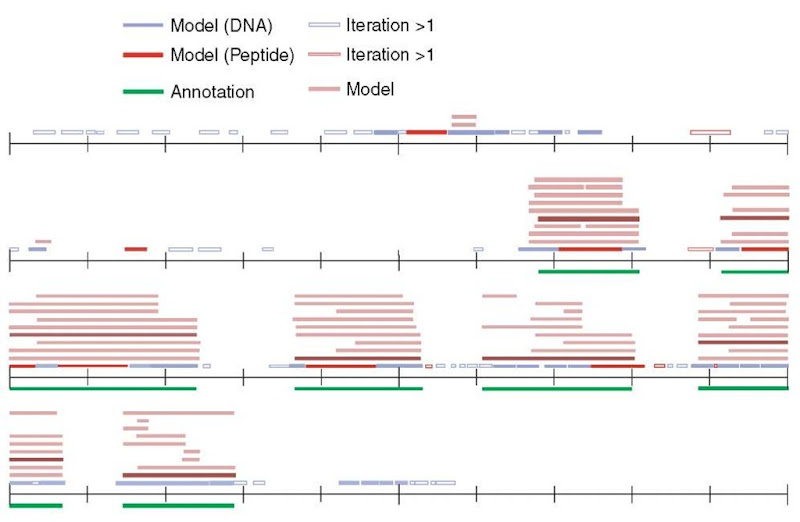
3. A candidate exon (CE) is defined as a region of local sequence similarity bounded by conserved splice sites or start/stop codons, respectively. Note that a region of local sequence conservation can be bounded by more than one conserved splice signal, so it can give rise to multiple overlapping CEs, as illustrated in Figure 1. Each CE is assigned a score, essentially depending on the level of sequence similarity and the quality of the splice signals.
Figure 1 Gene prediction by AGenDA. A human genomic sequence of 40 kb in length has been aligned to its counterpart in the murine genome (not shown in the figure). Green lines below the sequence are known protein-coding exons. Blue and red lines above the sequence correspond to regions of local sequence similarity as identified by DIALIGN. Pink lines correspond to candidate exons (CEs), that is, to local sequence similarities bounded by conserved splice sites or start/stop codons. A complete gene model is obtained as an optimal chain of CEs (purple lines). Local sequence conservation between human and mouse roughly corresponds to true exons, but is not very specific as many similarities outside the coding regions have been found. CEs bounded by conserved signals more accurately reflect the coding regions. Searching for optimal chains of CEs further reduces the noise of false-positive predictions; in our example, the optimal gene model identified by AGenDA exactly corresponds to the real gene
4. A candidate gene is a chain of CEs meeting some obvious formal conditions for splice signals and start/stop codons and certain length restrictions for introns. Candidate genes are considered on the forward as well as on the reverse strand.
5. The program uses a dynamic-programming algorithm to identify a set of candidate genes with maximum total score.
3. Results and discussion
AGenDA has been evaluated on standard reference sequences from human and mouse (Batzoglou et al., 2000). We found that the prediction quality of AGenDA is similar to the Hidden Markov Model (HMM)-based program GenScan (Burge and Karlin, 1997), which is currently the most popular software tool for gene prediction in eukaryotes. It is remarkable that a method solely based on evolutionary sequence conservation yields results that are comparable to the output of sophisticated intrinsic methods. While GenScan uses species-dependent statistical models to distinguish coding from noncoding regions, comparative methods are based on simple and universally applicable measures of local sequence similarity and on basic models for splice signals. Since these two gene-finding approaches are based on completely different types of input information, they complement each other and genes missed by one method can be detected by the other approach.
An obvious way of improving gene-prediction accuracy is to combine the predictive power of stochastical and comparative approaches. The effectiveness of such an integrated approach has been demonstrated by Korf et al. (2001) with their TWINSCAN program. In short, they reimplemented GenScan but also included homology information obtained from high-scoring BLAST alignments of syntenic sequences. This resulted in markedly increased sensitivity and specificity. Other combinations of intrinsic and comparative methods have been proposed by Meyer and Durbin (2002) and by Parra et al. (2003).
We are planning to integrate our alignment-based approach with the intrinsic gene-prediction program AUGUSTUS (Stanke and Waack, 2003). AUGUSTUS is based on a generalized HMM, with a new submodel for intron length distribution. With this new model, AUGUSTUS is superior to standard gene-finding programs if large input sequences are to be analyzed. Especially for genes with long introns, AUGUSTUS is far more accurate than other intrinsic methods. The stochastic model used by AUGUSTUS can incorporate external information in a natural way (Stanke, 2003). This way, we will use genomic alignments calculated by CHAOS and DIALIGN to further improve the performance of AUGUSTUS and to combine the advantages of these two approaches.