1. Introduction
Ever since the first genome sequence of Haemophilus influenzae was determined in 1995, there has been an explosion in the number of organisms whose genomes have been completely sequenced and made available in public databases. As on December 2004, there were 235 organisms whose complete genome sequences are available, including that of human (Bernal et al., 2001). Such an exponential increase in the number of available sequences has led to the realization that to understand the biology of an organism, one has to be able to make sense of this sequence data. One way of reaching this end is by identifying the function of proteins and RNAs encoded in its genome.
Traditionally, computational methods used to assign protein function are based on the simple assumption that proteins with similar sequences (homologs) perform similar molecular function. In such homology-based methods, function of a protein is inferred by comparing its sequence against a database such as the nonredundant database or Swiss-Prot using powerful tools such as PSI-BLAST (see Article 39, IMPALA/RPS-BLAST/PSI-BLAST in protein sequence analysis, Volume 7) to pick up homologs and then making good use of any available information on the homologous protein (Aravind and Koonin, 1999b).
Even though homology-based methods, such as those described above, have been quite effective at predicting molecular functions of remote homologs, large gaps still exist in our knowledge. Several genes in any given organism remain uncharacterized, where even sophisticated homology-based methods fail to suggest any function (Madera et al., 2004). Moreover, higher-order function of a protein, such as the pathway in which it plays a role, cannot, in principle, be assigned using such methods (Huynen et al., 2000). It is here that homology-independent methods, which primarily utilize the context in which a gene exists, become useful in inferring function. Such methods, more generally termed contextual methods, make use of information about gene organization in multiple genomes, and data obtained from large-scale functional genomics experiments such as gene expression, transcriptional interaction, and protein interaction studies.
In this article, we have classified homology-independent contextual methods to infer protein function into two major groups: methods that use genome sequence data and those that use information from large-scale functional genomics data.
2. Inferring function from genomic data
With a large number of genome sequences determined and many more in the pipeline, there is no paucity of data for gaining useful insights into protein function. In the following section, we discuss the different homology-independent contextual methods that use genome sequence data for inferring protein function.
2.1. Gene fusion
In a set of well-characterized proteins, it has been observed that pairs of interacting proteins or those that are functionally linked (e.g., involved in the same metabolic pathway) in one organism are fused into a single polypeptide chain in another organism. If two proteins are required to interact physically or are functionally linked, then it is likely that there is a selective pressure to produce them at the same time and at the same place within the cell, so that the chances that they interact are maximized. The best way to do it would be to have them as a part of the same gene product so that they are transcribed and translated at the same time and at the same place, thus allowing them to carry out their function efficiently. This logic has been exploited to predict pairs of interacting proteins by identifying instances of protein pairs that are fused in one organism but are found to be separate in another organism (see Figure 1a). A well-known example is the case where the genes encoding GyrA and GyrB in Escherichia coli are fused into a single-gene product, DNA topoisomerase, in yeast. Also illustrative is the case of gene fusions between evolutionarily mobile helicase and nuclease domains in DNA repair proteins, implying a general tendency of these domains to interact physically. An example is the fusion between PolIII subunit-like nuclease and DinG helicase in Bacillus subtilis (Aravind, 2000).
In one of the early computational analysis, Marcotte et al. (1999a) used genome sequence data and found that there are 6809 candidate interacting protein pairs in E. coli. The authors also used a three-step validation for their prediction: (1) ask whether the predicted pair of interacting proteins share a keyword between their annotations, which they show is very rare in randomly chosen pairs, (2) validate against experimental data, and (3) compare with results obtained from phyletic profiles (see Section 2.3 and Article 41, Phylogenetic profiling, Volume 7). Enright et al. (1999) showed that there are 215 proteins in the genomes of Escherichia coli, Methanococcus jannaschii, and Haemophilus influenzae, which are involved in 64 gene fusion events. In another similar study, it was shown that metabolic enzymes exhibited a profound 300% preference to exist as fused proteins over a control set of proteins (Tsoka and Ouzounis, 2000). This observation allowed them to conclude that proteins predicted to interact by detection of gene fusion events are more likely to be involved in metabolic pathways.
Figure 1 Figure illustrating inference of protein function from (a) gene fusion events and (b) conservation of gene order across multiple genomes
This method, while being useful, has a few pitfalls. In particular, this method could be confounded by evolutionarily mobile promiscuous domains that show fusions to other domains in a wide range of functional contexts. False-positives, with regard to prediction of physical interactions, may also crop up because it is perfectly possible that two fused protein domains do not interact physically but only functionally. The method cannot differentiate between interacting and noninteracting homologs. Another important problem associated with this method and the other genome sequence-based methods, which will be described later, is the difficulty in identifying true orthologs. If the stand-alone polypeptides and the individual components of the composite protein under consideration are really paralogs and not orthologs, we will end up with a false prediction (Marcotte et al., 1999a; Galperin and Koonin, 2000).
2.2. Gene order conservation and co-occurrence of genes in operons
Comparative genomic analyses among closely related species revealed an important fact that it is very rare for organisms to conserve gene order. Given the extremely high rate of recombination in bacterial genomes, the probability that the order of genes is maintained across many organisms is negligible. However, when gene order conservation is indeed observed, it only means that it is due to a selective pressure that the genes are kept together. This gene order conservation might mean that the two genes concerned encode products that either interact functionally or physically (see Figure 1b). In fact, Demerec and Hartman (1959) observed that the “mere existence of such arrangements shows that they must be beneficial, conferring an evolutionary advantage on individuals and populations which exhibit them” (Overbeek et al., 1999a). When interacting proteins are coded for by a polycistronic mRNA, the collection of adjacent genes is called an operon. Operons are common in prokaryotes and proteins involved in a particular biochemical pathway are generally clustered together to form an operon.
While this method is conceptually simple, in silico identification of operons in genome sequence is not trivial (see Article 20, Operon finding in bacteria, Volume 7). It was observed that the number of instances of functionally linked gene pairs increased with the increase in the number of organisms considered for the analysis. Overbeek et al. (1999a) have shown that the number of pairs of close bidirectional best hits (PCBBHs) identified in local alignment sequence searches is related to the square of the number of genomes considered. Data from 24 organisms pointed to the presence of 34 644 PCBBHs. More recent information on function inference from operons has shown that there are 58 498 PCBBHs in 31 genomes (Overbeek et al., 1999b). With the number of genome sequences being made available increasing exponentially, the predictive power of this method should increase at a much higher rate.
2.3. Correlated and anticorrelated occurrence of genes across many genomes
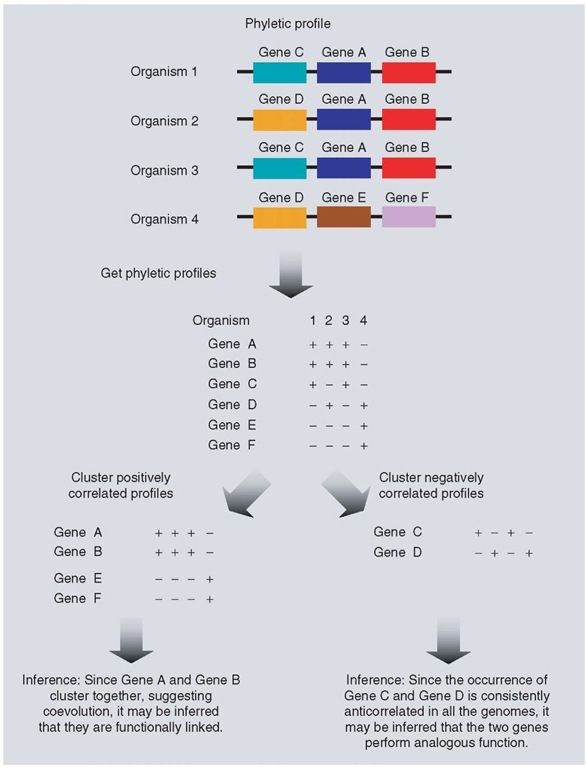
The idea behind this method described by Pellegrini et al. (1999) is that functionally related genes evolve in a correlated fashion across genomes. This implies that these genes should coexist across a set of organisms displaying the function of interest. In this technique, each gene is represented by a “phyletic profile” (see Article 41, Phylogenetic profiling, Volume 7), which is a string of n characters, where n is the number of organisms considered. Each of these characters may be either + or – representing the presence or absence of the gene of interest in the corresponding organism. The genes are then clustered into groups on the basis of their phyletic profiles. Gene products that belong to a cluster are then predicted to be functionally linked (see Figure 2).
In their study, Pellegrini et al. (1999) characterized 4217 genes from E. coli using this method. The number of organisms used to create the phyletic profile was 16. As an example, proteins with functions associated with the ribosome are discussed (Pellegrini et al., 1999). The ribosomal protein RL7 has homologs in 10 of the 11 eubacterial genomes and in yeast, but not in archaea. More than 50% of E. coli genes with function associated with the ribosome share the phyletic profile with RL7. Several uncharacterized proteins were found to fall under this cluster, and the authors state that it is likely that these uncharacterized proteins have functions associated with the ribosome. These proteins whose function has been inferred by this method share no sequence similarity to the characterized protein such as RL7, and hence their role in the functioning of the ribosome could not have been inferred by homology-based methods. Another example of application of this method to inferring function is the subcellular localization of proteins. It is based on the idea that proteins that localize to a particular cellular compartment have a similar phyletic profile. This method, when applied to Saccharomyces cerevisiae identified 361 nucleus-encoded mitochondrial proteins with 50% accuracy and 58% coverage (Marcotte et al., 2000).
Figure 2 Inference of protein function using correlated or anticorrelated phyletic profiles of clusters of genes. “+” represents presence of the gene and “-” represents absence of the gene
Just like how similarity in phyletic profiles of genes can be used to infer the function of a protein, anticorrelation of phyletic profiles can also be used to identify instances of nonorthologous gene displacement, that is, instances in which nonhomologous proteins perform the same function in different organisms. The principle behind this method relies on identifying cases in which two genes have dissimilar phyletic profiles, that is, if the first gene is present in an organism, then the second gene is absent in the same organism. If this observation is consistent for a large number of organisms considered, then one can infer that the two proteins perform analogous function and evolution has selected for one of the two proteins, discarding the other (see Figure 2).
An example of how anticorrelated phylogenetic profiles have enabled us to understand a well-characterized metabolic pathway such as glycolysis involves the enzyme fructose-1,6-bisphosphate aldolase (FBA). This enzyme catalyzes the step where fructose-1,6-bisphosphate is cleaved to glycerol-3-phosphate and dihydroxy-acetone phosphate. While this enzyme is present in most bacteria and eukaryotes, it is absent in Chlamydiae and Archaea. However, a second enzyme DhnA in these organisms forms an almost-perfect complementary phyletic pattern against a set of reference genomes, indicating that this enzyme is the only FBA in Chlamydiae and Archaea (Galperin and Koonin, 2000). In another case, seven enzymes belonging to the 15-step thiamin biosynthesis pathway were found to have been displaced by analogous proteins. These predictions have been verified experimentally. Importantly, this led to the assignment of function for three proteins, till then uncharacterized. An example is the displacement of thiamin phosphate synthase, first described in E. coli, by genes orthologous to the hypothetical ORF MTH861 in M. thermoautotropicum (Morett et al., 2003).
2.4. Conservation of bidirectionally transcribed gene pairs
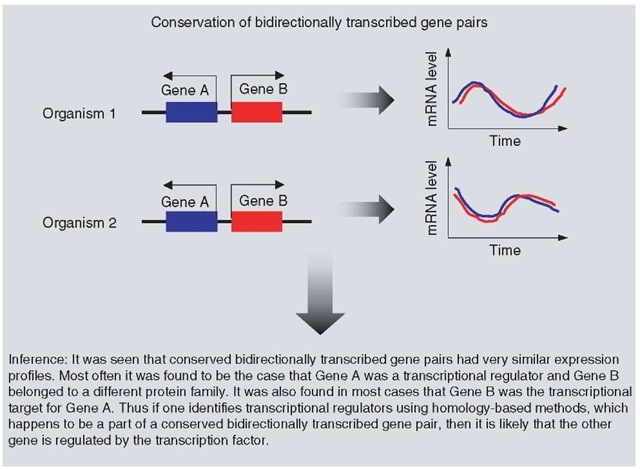
Yet another contextual method for inferring function from genomic data is the identification of conservation of gene orientation of adjacent, bidirectionally transcribed gene pairs across genomes (Korbel et al., 2004). These bidirectionally transcribed gene pairs may be regulated by a pair of overlapping promoter elements. Such gene organization is beneficial as it offers a method of transcriptional regulation distinct from operons (Korbel et al., 2004; Warren and ten Wolde, 2004). It is also worthwhile to note that while organization of convergently transcribed gene pairs is rapidly lost during evolution that of divergently transcribed gene pairs is not. Systematic analysis carried out by Korbel et al. (2004) shows that over 5000 divergently transcribed gene pairs are conserved across distantly related organisms and that 26% of all E. coli genes are divergently transcribed pairs, a quarter of which are conserved in a distant evolutionary clade. There are 6.5 times more divergently transcribed gene pairs than convergently transcribed ones. Such conservation of bidirectionally transcribed gene pairs may be due to the pressure of coexpression, similar to the pressure that maintains the operon structure. About 87% of gene pairs conserved across distant genomes have correlated gene expression, with a Pearson correlation coefficient value greater than 0.6 (see Figure 3).
Bidirectionally transcribed gene pairs may more than just be coexpressed. Over 71% of such pairs identified in E. coli were classified as “RX” by the authors, where one gene product is a transcriptional regulator (R) and the other belongs to some other class of protein (X). Interestingly, most of such RX pairs have been shown to be autoregulatory. Thus, if a protein has been classified as a transcription factor by homology-based methods (Aravind and Koonin, 1999a; Perez-Rueda and Collado-Vides, 2000; Madan Babu and Teichmann, 2003), then its transcriptional target in vivo can be determined using this method. For example, using this method, it was shown that prokaryotic members belonging to the orthologous group KOG2969 (see Article 90, COGs: an evolutionary classification of genes and proteins from sequenced genomes, Volume 6) are involved in regulating the expression of ribosome-associated genes in alpha-proteobacteria. It has been experimentally shown that KOG2926, whose gene is divergently transcribed from the nearby mito-chondrial ribosomal protein S2 (RpsU) gene, is indeed the transcriptional regulator of rpsU. The efficiency of this method, like that of any other genomic context method, is dependent on the number of genomes sampled (Korbel et al., 2004).
Figure 3 Determining the function of proteins from conservation of bidirectionally transcribed gene pairs
3. Inferring function from large-scale experimental data
Coupled with the explosion in the availability of complete genome sequence for several organisms is public access to large-scale experimental data on interactions between the cellular components and to data on gene expression for several organisms obtained using microarrays. These interactions may be protein-protein, protein – metabolite, or protein-nucleic acid.
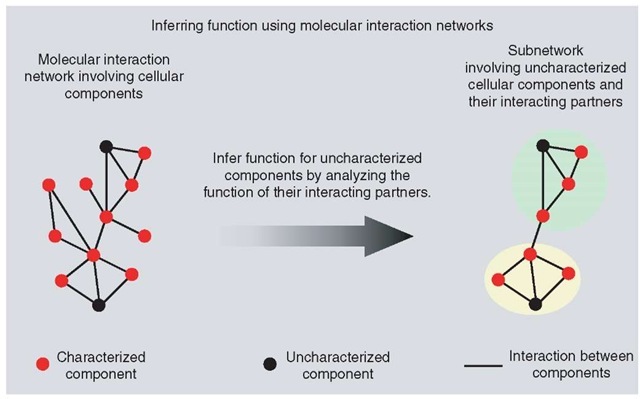
The following section will focus on methods that utilize data on the interactions between the cellular components and gene-expression studies to infer functions of uncharacterized genes. The set of all interactions mediated by cellular components can be conceptualized as a graph or a network, in which each cellular component is represented as node and interaction between the two components as edges or arcs. Such cellular networks can be used to infer protein function in the context of a biological process (Barabasi and Oltvai, 2004; Xia et al., 2004). For instance, a link between a functionally uncharacterized protein X and well-characterized protein(s) will enable one to place the function of X in context of the function of the characterized protein(s) to which it is linked (see Figure 4).
In the following section, we review four different methods of inferring protein function using data obtained from the different large-scale experiments.
Figure 4 Figure illustrating the general idea of inferring function of an uncharacterised cellular component based on its links with other characterized components in a molecular interaction network
3.1. Protein-protein interaction network
Identification of cellular protein-protein interaction networks has become possible due to the development of high-throughput techniques such as the yeast two-hybrid experiment and the tap-tag method (see Article 37, Inferring gene function and biochemical networks from protein interactions, Volume 5). Protein-protein interaction networks derived using the two-hybrid method are available for D. melanogaster (Giot et al., 2003), C. elegans (Walhout et al., 2000), S. cerevisiae (Uetz et al., 2000), H. pylori (Rain et al., 2001), and the T7 bacteriophage (Bartel et al., 1996). Such experimentally derived networks can be analyzed using computational approaches and useful functional information can be obtained from them.
When a protein-protein interaction network is rendered in such a way that specific functional categories are highlighted, it will be observed that proteins belonging to the same functional category cluster together. It has been shown that 72% of interactions between experimentally characterized proteins in such networks are between pairs belonging to the same functional class. The significance of this number is illuminated when we see that this percentage is only 12% when the network is randomly divided into different categories. Linkages between proteins belonging to different functional categories are also possible. The difficulty with interpreting such linkages is that they may be false-positives or genuine cross talk between related pathways. It was also determined that 78% of interactions between proteins with known cellular localization involved proteins sharing at least one cellular compartment (Schwikowski et al., 2000; Tucker et al., 2001).
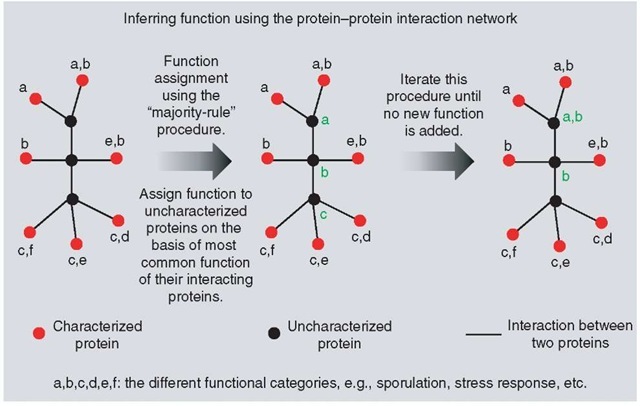
Protein-protein interaction networks have been used to assign functions to a number of previously uncharacterized proteins. A common approach for assigning function using protein-protein interaction network is what is known as the majority-rule assignment. This technique assigns function on the basis of most common functions present among the characterized interacting partners (see Figure 5). Vazquez et al. (2003) have used interactions between uncharacterized proteins in an iterative majority-rule technique to yeast and identified 2238 interactions among 1826 proteins (Vazquez et al., 2003).
Map of 957 interactions involving 1004 yeast proteins generated by Uetz et al. (2000) has yielded novel insights into functions of several proteins. For instance, two yeast proteins of unknown function, which were seen to interact with each other, also bind to ornithine aminotransferase. This implies that they may be involved in arginine metabolism. Novel interactions between proteins involved in the same biological function were also shown. Their data also shows that novel interactions exist between proteins involved in diverse biological processes. For example, three snRNPs were found linked to the ribosomal protein S28. This might indicate a role for S28 in RNA splicing or, alternatively, a novel role of snRNPs in translation or ribosome biosynthesis (Uetz et al., 2000).
This method, like any other, is not free from operational difficulties and errors. The yeast two-hybrid method can only detect binary interactions, but in reality many proteins function as protein complexes. This may partly be overcome by the tap-tag method, which is used to purify protein complexes and characterize them. Even in such cases, interactions between the individual subunits of the complexes are not known. Yeast two-hybrid also gives rise to many false-positive interactions because of the nature of the experiment – since the interaction is tested in the nucleus, the local cellular environment may be different and it is possible that the interaction is mediated by a third protein, in which case the two proteins shown to interact by the two-hybrid system do not really interact directly (Aloy and Russell, 2002; von Mering et al., 2002).
Figure 5 Figure demonstrating the use of majority-rule assignment (see text) of function from protein-protein interaction networks. In this figure, labels a-f in black represent arbitrary functional class assigned to well-characterized proteins (nodes) in the network. Labels a-c in green are the functions of uncharacterized proteins (nodes) inferred using the majority-rule method
3.2. Transcriptional regulatory network
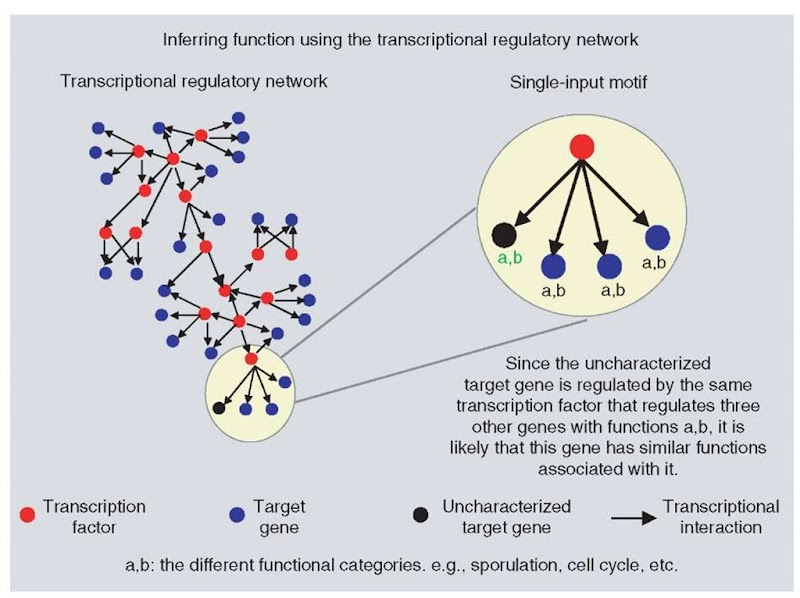
Transcriptional regulatory networks are deciphered by carrying out ChIP-chip experiments, which specifically identify DNA sequences that can interact with transcription factors. Data generated are called location data, and are again useful in assigning function to proteins (see Figure 6).
A transcriptional regulatory network has been generated by Lee et al. (2002) for yeast. This study used 141 transcription factors (TFs) that were listed at that time in the Yeast Proteome Database. Out of these 141 TFs, only 106 gave useful information, as experimental modification caused loss of function in 17 proteins and the others could simply not be detected. This network showed that 2343 of 6270 yeast genes were bound by one or more of the 106 TFs when the cells were grown in rich media. More than one-third of these promoters were bound by more than one TF. In the assembled network, six network motifs were identified. Network motifs are short patterns of interconnections that recur at many places in the network at frequencies much higher than seen in random networks of a similar size (Lee et al., 2002; Shen-Orr et al., 2002). In the yeast network, the motifs identified were the following: autoregulation, multicomponent loops, single input, multi-input, feed-forward loops, and regulator chain. Since each network motif has a specific information-processing task (Shen-Orr et al., 2002), such regulatory motifs could be used to make functional assignments. For example, Fhl1, a protein with function unknown was found to be involved in a single-input motif-regulating multiple genes involved in ribosome biosynthesis. This protein was also involved in a multi-input motif. Experiments have showed that a mutation in this protein caused serious defects in the ribosome synthesis machinery (Lee et al., 2002).
Figure 6 Determination of protein function using the context in which it occurs in the transcriptional regulatory network
Knowledge about the function of proteins could be extended using such networks. For example, the protein Phd1 that is involved in pseudohyphal growth in nutrient stress conditions was shown to interact with proteins involved in general stress response and in regulation of metabolism (Lee et al., 2002).
More recent transcriptional regulatory network generated for yeast by Harbison et al. (2004) has revealed 11 000 interactions mediated by 203 TFs. In this study, predictions of TF-promoter pairs were validated using comparisons across four species of yeast. This work discovered and rediscovered promoter elements. Functional correlations, to confirm previous functional assignments, could be made to the network connections. For example, six cell-cycle transcriptional regulators were found to bind to the promoter for YHP1, which is involved in the regulation of the G1 phase of the cell cycle.
3.3. Gene coexpression network
Since proteins involved in the same pathway or those that are part of the same protein complex are coregulated at the transcriptional level, gene-expression data can be used to make functional inferences. However, coexpression alone will not necessarily mean functional relationship (Mateos et al., 2002). On the other hand, when a pair of gene is consistently coexpressed across large evolutionary distances, this method becomes a powerful tool to infer protein function (Teichmann and Madan Babu, 2002; Stuart et al., 2003; van Noort et al., 2003; McCarroll et al., 2004) (see Figure 7).
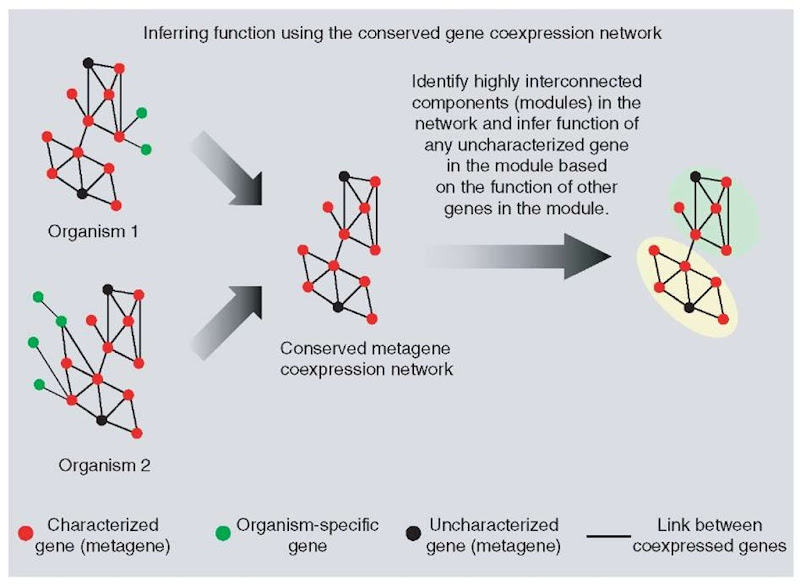
Stuart et al. (2003) have constructed a gene coexpression network (see Article 60, Extracting networks from expression data, Volume 7) for a set of metagenes. Metagenes are defined as sets of genes that are evolutionarily conserved across diverse organisms (effectively the term metagene is identical to a group of orthologous genes). In such a network, metagenes are represented as nodes and gene coexpression between a pair of genes as links. In their work, they first characterized 6307 metagenes using the following organisms: human, fly, worm, and yeast. For each pair of metagenes, coexpression was studied and gene pairs whose expression was significantly correlated across multiple organisms were identified. Expression was studied only for metagenes because the evolutionary conservation of these genes implies that the network covers only core biological processes. This coexpression network comprised 3416 metagenes connected by 22 163 interactions. As a next step, the authors used a visualization technique in which metagenes were placed next to each other at a distance in proportion to their level of coexpression. Highly connected and conserved regions in the network could be visualized as peaks in the map. Such visualization led to the identification of 12 “components” -regions containing a large number of interconnected metagenes. These components were the following: signaling, ribosome biogenesis, energy generation, proteasome, cell cycle, general transcription, animal specific, translation, ribosomal subunits, secretion, neuronal, and lipid metabolism (Stuart et al., 2003).
Figure 7 Use of conserved gene coexpression networks to determine protein function. The method initially identifies metagenes, which are groups of orthologous proteins across the different species. For each organism, a gene coexpression network is determined and the part of the network that is conserved in the set of organisms considered is referred to as the conserved metagene coexpression network
The function of three uncharacterized genes was determined using this method. It was found that these proteins were coexpressed with genes involved in cell proliferation and cell cycle. Two other proteins known to function in other cellular processes also coexpressed with the above set of metagenes. Experimental verification of these results showed that all these five proteins were overexpressed in human pancreatic cancers relative to normal tissue and hence are involved in cell proliferation. Thus, by analyzing such coexpression networks, one can identify previously unknown proteins involved in well-characterized biological processes.
3.4. Integration of different data types
The methods discussed in the previous sections make use of a single type of data to infer protein function. The next logical step is to integrate different types of data to achieve this objective.
Marcotte et al. (1999b) have integrated data from phyletic profiles, gene fusion, and gene expression with the experimentally determined protein-protein interaction networks in yeast and identified 4130 links of very high confidence. They use this network to assign function to proteins. The yeast ORF, YGR021W, was assigned a function related to mitochondrial protein synthesis. The function of Sup35 was extended to beyond its role as a translation release factor and in guiding nascent proteins to their cellular locations. Sup35 shows both correlated evolution and expression pattern with a yeast chaperonin system thought to aid in the folding of newly synthesized actin and microtubules.
Bar-Joseph et al. (2003) have combined gene-expression data and location analysis data to assign functions to proteins. They have done this because they realized that location analysis, while identifying what interacts with what, does not throw light on the type of interaction, that is, whether the interaction activates or represses transcription. To infer this, it is best to use gene-expression data. The algorithm that they have developed, namely, GRAM (Genetic Regulatory Modules), clusters sets of genes to which a common TF is bound. From this set, a subset of genes with correlated expression is derived. Positive correlation between TF binding and expression level indicates that the regulator protein is an activator. The integration of the two types of data allows a relaxation of the conditions set to identify a binding event. This will bring down the number of false-negatives while not substantially increasing the rate of false-positives. This reduction in stringency allowed six new genes to be identified as targets of Hap4, a well-characterized protein involved in regulation of oxidative phosphorylation and respiration.
In another recent work, Luscombe et al. (2004) developed an approach called SANDY (Statistical Analysis of Network Dynamics), which integrates gene-expression data with the transcriptional regulatory network to identify condition-specific transcriptional regulatory networks. By integrating these two sources of information, novel insights about the dynamic nature of transcriptional regulatory networks were obtained. For instance, this approach led them to identify “master regulators” that are important in particular cellular processes, such as sporulation and cell cycle. This approach additionally helped in identifying condition-specific transcription factors and their transcriptional targets for a given cellular condition such as stress and DNA damage.
4. Conclusions
The above, recently developed methods for protein function prediction have created a paradigm shift in our perspective toward understanding proteins. The classical view of protein function in terms of its molecular properties such as catalysis or ligand binding is being complemented by what is known as contextual or cellular function (see Article 29, In silico approaches to functional analysis of proteins, Volume 7). It is defined as the understanding of protein function in terms of the pathways or intracellular subnetworks in which the protein might be expected to play a role (Fraser and Marcotte, 2004; Palsson, 2004). The actual role it plays in this context would need to be elucidated by further experimentation. As in the case of homology-based methods, it is again due to the availability of a large volume of organized data that nonhomology-based methods to infer protein function have become feasible. Such large-scale methods for predicting protein function becomes more important in light of the situation where over a thousand genes in the most characterized bacterial genome, E. coli, remain known only as hypothetical ORFs.