1. Introduction
Python is an interpreted, interactive, object-orientated programming language first created and released by Guido van Rossen in 1989-1991. It is called Python after the BBC comedy series, “Monty Pythons’s Flying Circus”. Python is currently an open source software project, which freely welcomes anyone with an interest in the language to contribute to it. The primary source of information concerning Python can be found at www.python.org, which includes downloads of the most recent release of the language (2.3.4) for all major operating systems. Other references to bioinformatic scripting in general and Python in particular can be found in this topic in other article references (see Article 103, Using the Python programming language for bioinformatics, Volume 8, Article 112, A brief Perl tutorial for bioinformatics, Volume 8, and Article 104, Perl in bioinformatics, Volume 8).
2. Language basics
Python uses a natural language syntax and indentation to delineate code blocks (as opposed to {} brackets used by most other programming languages). Control statements are terminated by a colon. Python is case sensitive, x and X could be the names of different variables. Python is a dynamically/implicitly typed language, which means that variables do not need to be formally declared at compile time. Variables can also be reused to hold any type of data during the runtime of the program. The Python interpreter can be run in interactive mode, either from a Unix style command line or by running pythonwin in Win32. This shell is exited with control-D. Inside the interpreter, lines are prefixed by “>>>”. A Python script can be run by passing it as a parameter to the Python command. The -i parameter will invoke the interactive interpreter after script execution.
For example, assume that the script myscript.py contains the line x = 1:
Basic data types: Python has four basic data types: numbers, strings, 0-based arrays, and dictionaries (hash tables). Literal strings can be enclosed by either single or double quotes. There are two types of arrays: mutable lists (contents can be changed after instantiation) and immutable tuples.
Assignment uses the = operator:
3. Basic data manipulation
Strings and arrays can be sliced, which means you can extract the nth element or the m th to n th element.
Strings can be instantiated with % substitution
File handlers can be assigned using the built-in open function. Files can be opened in read (r), write (w), append (a), and binary (b) modes. r is the default mode.
Variables can be assigned a null/nil value with the keyword None.
Boolean operators: Python uses natural language : and, or, not, in.
Equality operators: == (equals), != (not equals), < (less than), > (greater than).
Any variable can be used in a boolean statement. None, False, “”, 0, [], and () equate to false, any other value equates to true. The boolean types True and False can also be used.
4. Importing modules
External Python modules are imported using the import key word. Modules can be any other Python scripts written by anyone, defined modules that come with the language, or third-party modules downloaded and compiled into the Python program. Paths to Python modules in the first case can be defined either with the environment variable PYTHONPATH or at runtime using the path function of the sys module (see below). Import statements can occur anywhere in Python code (including inside if … else statements). There are two syntaxes for the import statement, which affect how members of the module are referenced. The following example assumes a file called mymodule.py, which is located in mydi-rectory/mypythonscripts.
5. Control statements
Python uses if … elif … else, for and while for execution control.
While and for loops can be exited with the break statement.
Python uses class to define a new class, and def to define a function/ subroutine or class method. Function parameters can be assigned a default value, and those parameters with default values are optionally passed to the function when called.
Class syntax is shown in the examples below. Python has no concept of class scope, such as private or protected, as found in languages such as Java. It is common to use single or double underscore prefixes as a “reminder” that a method has restricted scope, but this rule is not enforced by Python. You can overload built-in functions using methods both prefixed and suffixed by double underscores; examples are given below. Class methods always receive the keyword self as the first parameter; class attributes and methods are also referenced within the class by the self keyword.
Python is inherently reflective. Variables and functions can be accessed through the built-in locals() and globals() functions, which return dictionaries of all elements of the script. Classes can be manipulated with the hasattr, getattr, and setattr built-in methods. Examples of these are below.
6. Python bioinformatic resources
There is an open source project located at www.biopython.org, which contains a large number of script and class modules to handle biological data and which acts as a framework for building your own programs. A short example is shown in the examples section.
7. Examples
The following example scripts display the flexibility of the Python language for building useful code for biological data manipulation. The code can be cut and pasted into files, as indicated.
Running the biorun script (Python biorun.py from the command line) will produce the following output (from the print statements):
It will also produce a file called “sequences.txt,” which contains the following:
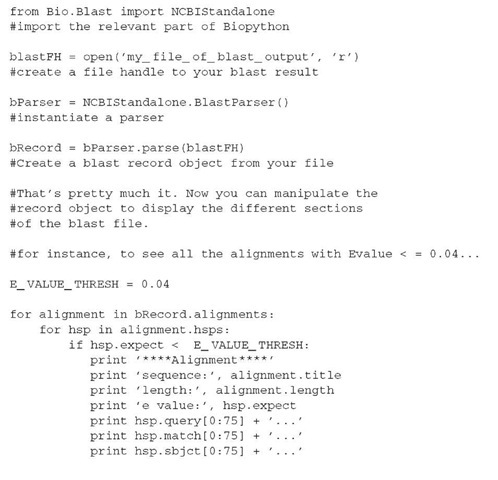
8. Biopython example
The following is paraphrased from www.biopython.org/docs/tutorial/Tutorial004.html
Assuming that you have downloaded and installed the Biopython modules from www.biopython.org and that you have a local copy of a blast search,
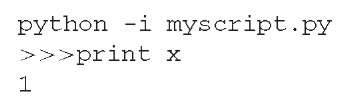










![tmp1A5-81_thumb[1] tmp1A5-81_thumb[1]](http://lh3.ggpht.com/_NNjxeW9ewEc/TJxKwyAD-hI/AAAAAAAAERg/yLRjR6rhR14/tmp1A581_thumb1_thumb1.jpg?imgmax=800)
![tmp1A5-82_thumb[1] tmp1A5-82_thumb[1]](http://lh6.ggpht.com/_NNjxeW9ewEc/TJxKymCnwII/AAAAAAAAERo/6B5WaBlC2_E/tmp1A582_thumb1_thumb1.jpg?imgmax=800)
![tmp1A5-83_thumb[1] tmp1A5-83_thumb[1]](http://lh6.ggpht.com/_NNjxeW9ewEc/TJxK1xIGTqI/AAAAAAAAERw/iN04Ruj2GsQ/tmp1A583_thumb1_thumb1.jpg?imgmax=800)