1. Introduction
The article “RNA secondary structure prediction” discussed dynamic programming methods to predict the minimum free energy (mfe) E0 and minimum free energy secondary structure S 0 of a given RNA sequence, using the Turner energy model (Xia et al., 1999), with experimentally measured negative, stabilizing base stacking energies and positive, destabilizing loop energies (hairpin loop, interior loop, etc.). Here, we survey a few applications of this method to determine regulatory regions of RNA and more generally to determine noncoding RNA genes.
2. Methods
A general, often-used approach in genomic motif finding is to fix a window size n, and scan through a chromosome or genome, repeatedly moving the window forward one position. The window contents may then be scored using machine-learning algorithms, such as weight matrices (Gribskov et al., 1987; Bucher, 1990), hidden Markov models (Baldi et al., 1994; Eddy et al., 1995; see also Article 98, Hidden Markov models and neural networks, Volume 8), neural networks (Nielsen et al., 1997; see also Article 98, Hidden Markov models and neural networks, Volume 8), and support vector machines (Vert, 2002; see also Article 110, Support vector machine software, Volume 8). While accurate detection of protein coding genes can be achieved using hidden Markov models (Borodovsky and McIninch, 1993; Burge and Karlin, 1997), by exploiting the nucleotide bias present in a succession of codons, such signals are less apparent in noncoding RNA genes.
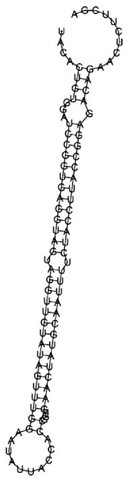
Noncoding RNA (ncRNA) (Eddy, 2001; Eddy, 2002) is transcribed from genomic DNA and plays a biologically important role, although it is not translated into protein. Examples include tRNA, rRNA, XIST (which in mammalian males suppresses expression of genes on the X chromosome) (Brown etal., 1992), metabolite-sensing mRNAs, called riboswitches, discovered to interact with small ligands and up- or downregulate certain genes (Barrick et al., 2004), tiny noncoding RNA (tncRNA) (Ambros et al., 2003), and miRNA (microRNA). MicroRNAs are ~21 nucleotide (nt) sequences, which are processed from a stem-loop precursor by Dicer (Tuschl, 2003; Lim et al., 2003) – see Figure 1, which depicts the predicted secondary structure for C. elegans let-7 precursor RNA. MicroRNA is (approximately) the reverse complement of a portion of transcribed mRNA, and has been shown to prevent the translation of protein from mRNA – this is an example of posttranscriptional regulation.
Figure 1 Predicted minimum free energy secondary structure of C. elegans let-7 precursor RNA; sequence taken from Rfam. Predicted minimum free energy for this 99-nt sequence is -42.90 kcal mol-1 (prediction made using Vienna RNA package)
For certain classes of ncRNA, there is a sufficiently well-defined sequence consensus or common secondary structure shared by experimentally determined examples, so that machine-learning methods such as stochastic context-free grammars (SCFG) have proven successful. RNA secondary structures can be depicted as a balanced parenthesis expression with dots, where balanced left and right parentheses correspond to base pairs and dots to unpaired bases.
In particular, by training an SCFG on many examples of tRNA, additionally using promoter detection with heuristics, T. Lowe and S. Eddy’s program tRNAscan-SE identifies “99-100% of transfer RNA genes in DNA sequence while giving less than one false-positive per 15 gigabases” (Lowe and Eddy, 1997).
Exploiting the fact that ncRNA genes of the AT-rich thermophiles Methanococ-cus jannaschii and Pyrococcus furiosus have high G + C content, Klein et al. (2002) describe a surprisingly simple yet accurate noncoding RNA gene finder for these and related bacteria. Lim et al. (2003) describe a novel computational procedure, MiRscan, to identify vertebrate microRNA genes. In a moving-window scan of the noncoding portion of the human genome, MiRscan uses RNAfold from the Vienna RNA Package (Hofacker et al., 1994) to search for stem-loop structures having at least 25 bp and predicted mfe of -25 kcal mol-1 or less. Subsequently, MiRscan passes a 21-nt window over each conserved stem-loop, then assigns a log-likelihood score to each window to determine how well its attributes resemble those of certain experimentally verified miRNAs of Caenorhabditis elegans and Caenorhabditis briggsae homologs.
Using the power of comparative genomics (alignments of homologous ncRNA genes from different organisms), Rivas and Eddy (2001) developed the program QRNA that trains a pair stochastic context-free grammar, given pairs of homologous ncRNA genes. Coventry et al. (2004) developed the algorithm MSARI, which assigns appropriate weights for local shifts of a ClustalW multiple sequence alignment of many (e.g., 11) homologous ncRNAs, in order to detect a conserved pattern of secondary structure. The authors suggest that a gene finder might then be trained on automatically generated multiple sequence alignments of RNAs, suitably corrected by their algorithm to identify the underlying sequence/structure alignment.
A related and equally important algorithmic task is the detection of regulatory and retranslation signals in the untranslated region (UTR), both upstream 5′ and downstream 3′ of the coding sequence (cds) of messenger RNA. For instance, Lescure et al. (1999) used Vienna RNA Package RNAfold in a simple screen to determine putative selenocysteine insertion sequence (SECIS) elements (see Huttenhofer and Bock, 1998 for a review of selenocysteine incorporation); the authors subsequently performed (wet-bench) experiments to validate certain SECIS elements. Grate (1998) applied Eddy’s RNA structure pattern searching algorithm program RNABOB in the search for SECIS elements in HIV. Bekaert et al. (2003) developed a model for -1 eukaryotic ribosomal frame-shifting sites, on the basis of a slippery sequence and a predicted pseudo-knot structure.
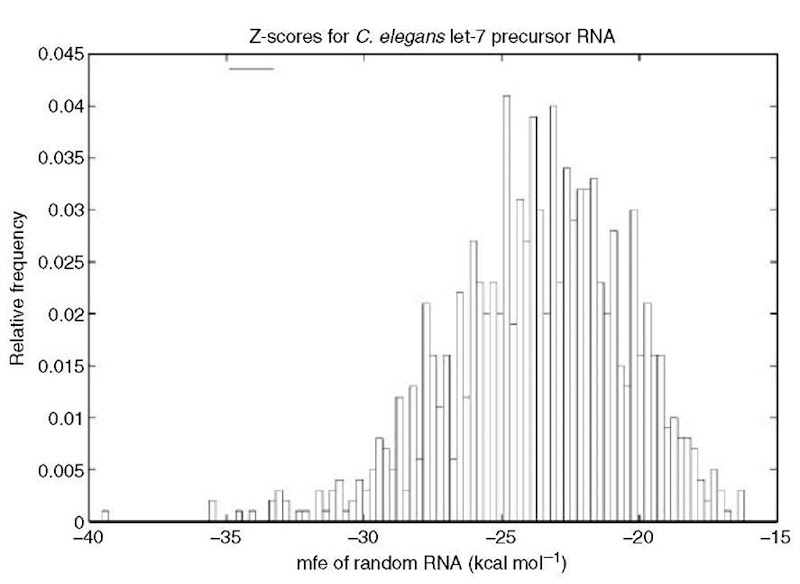
Recently, Washietl et al. (2005) described a noncoding RNA gene finder, based on a combination of mfe Z-score computations and comparative genomics. Here, the Z-score of the content of a current window of size n is defined by , where x is the mfe of the window contents, while a are respectively the mean and standard deviation of the mfe of random length n sequences having the same mono- or possibly dinucleotide frequencies as that of the window contents (see Workman and Krogh, 1999; Clote et al., 2005 for discussion, and Figure 2 for an example). A Z-score of x that is approximately zero means that the mfe of sequence x is indistinguishable from that of its randomizations (i.e., the mfe of a randomization of x is just as often lower as higher than that of x ). Similarly, a negative Z-score of x means that the mfe of x is lower than that of most of its randomizations.
Figure 2 Histogram of the mfe for 1000 random RNAs, each having the same (exact) dinucleotide frequency as that in C. elegans let-7 precursor RNA. Mean mfe is -23.54 kcal mol-1 with standard deviation 3.23, hence the Z-score for let-7 precursor RNA is -42’90-2-23’54) or roughly -6. Random RNA produced by the method of Workman and Krogh (1999) as implemented in Clote et al. (2005) (minimum free energy computed using RNAfold)
Results from Rivas and Eddy (2000) indicate that using Z-score alone is not sufficiently statistically significant to be used to find ncRNA genes. Nevertheless, Washietl et al. (2005) combine the use of Z-scores with comparative genomics to develop a remarkably accurate and computationally efficient noncoding RNA gene finder. The authors make novel use of a support vector machine to compute the mean \x and standard deviation a, rather than relying on slow repeated randomizations of window contents.