Aging research throughout the first three epochs of gerontology was primarily concerned with describing general aspects of the process covering all levels of biological organization, from the molecular to the organismal. The data collected spawned a large number of theories touching on all aspects of cellular structure and function, as well as changes that may occur at the physiological level. Although these theories were crucial for producing advances in the discipline, they failed to produce a clear picture of fundamental mechanisms responsible for the aging process. Gerontology was placed on firmer ground with an NIA program to isolate genes that influence longevity, an effort that has greatly improved the genetic analysis of the aging process.
Thus, with the beginning of the current epoch and the launching of comprehensive genome sequencing projects, the goal of gerontology shifted to the identification and characterization of genes that promote longevity. Despite their name, longevity genes were not always selected by evolutionary forces to give an organism a long life span. Quite the contrary, since some of these genes, when functioning normally, limit the life span; only after being mutated and made dysfunctional do they increase the organism’s life span. This type of longevity gene is said to be a negative regulator of life span because their normal function is to limit an organism’s life span. Other longevity genes are said to be positive regulators because expression (or overexpression) of these genes increases the life span.
The normal life span of an organism is produced by a complex mix of positive and negative regulator genes that seem to produce the optimum—not necessarily the longest—life span that best fits the organism’s size, metabolic rate, and activity level, as well as its position in the grander theater of predator-prey relationships. The search for longevity genes in yeast, nematode, Drosophila, mice, and humans has led to a much clearer picture of the mechanisms controlling the aging process. It has also shed light on how those mechanisms can be modulated to fine-tune an organism’s life span to maximize the survival, not of the individual, but of the species to which it belongs. But gerontologists expect that a clear understanding of all longevity genes will provide a way of reversing or forestalling human aging.
Yeast
Yeast are unicellular organisms that divide at regular intervals and, as a population, are nearly immortal. Each cell begins as a mother cell that produces a daughter cell each time it divides, but the mother cell ages with each cell division; thus its life span is limited to a finite number of cell divisions, after which it dies, while the daughter cells continue on for the same finite number cell divisions. The measure of the yeast life span is thus the number of divisions of the mother cell before it dies, not the amount time that it has lived. The identification of longevity genes in yeast provided the first comprehensive list consisting of four processes that are believed to control the aging process. These processes are metabolic control, resistance to stress, gene disregulation, and the maintenance of genetic stability.
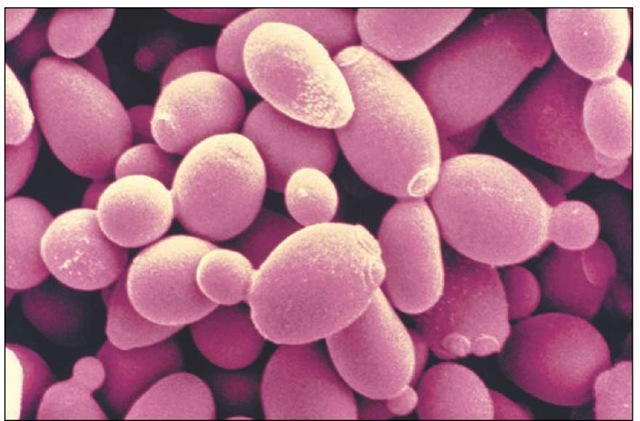
Yeasts, such as Saccharomyces cerevisiae, have been used by researchers in the search for longevity genes. This image shows several of the cells in the process of cell division by budding, which produces a daughter cell that is initially smaller than the mother cell.
The first longevity gene, called Lag-1 (longevity assurance gene number 1), was isolated from yeast by Dr. S. Michel Jazwinski and his team at Louisiana State University in 1994. Since that time, 14 additional longevity genes have been identified in yeast. The Lag-1 protein (Lag-1) is located in the membrane of the endoplasmic reticulum and is involved in the production of glycolipids.Glycolipids are an important component of the glycocalyx, a molecular "forest" that covers the surface of all cells. The glycocalyx is essential for cell-to-cell communication and contains many receptors that regulate a host of cellular functions. Many glycolipids are involved in signaling pathways that regulate growth, stress resistance, and apoptosis. Lag-1 is a positive regulator of life span, and while the mechanism by which it influences life span is unclear, a mutation in this gene could reduce the cell’s ability to cope with stress, to block proliferation, or to induce apoptosis.
All eukaryotes have an intracellular signaling pathway, known as the retrograde response, that serves to coordinate mitochondrial function with the expression of mitochondrial genes in the cell nucleus. Although mitochondria have their own genome, most of the Krebs cycle enzymes (all of which function inside the mitochondrion) are coded for by the cellular genome. The rate at which these genes are transcribed depends on how badly the mitochondria need the enzymes. During periods of stress, caused by high temperatures or an unfavorable environment, mitochondria are extremely active. Enzymes usually have a short life span, and during periods of extreme activity they must be replaced more frequently. The main function of the retrograde response is to ensure that the mitochondria always have enough Krebs cycle enzymes. Two other longevity genes, called Ras-1 and Ras-2 (rhymes with "gas"), regulate this pathway. Mutations in either or both of these genes eliminate the retrograde response, thus abolishing the cell’s ability to deal with stress of the kind described. Consequently, the cell does not receive sufficient amounts of ATP, the main energy source, at a time when it needs it the most, resulting in cellular damage and early death. Overexpression of Ras-2 can completely abolish the negative effect on life span of chronic heat stress. Yeast demonstrating natural thermotolerance early in life invariably have longer life spans than is normal.
Gene disregulation has been observed in yeast that lose transcrip-tional silencing of genes in heterochromatic regions of the genome (i.e., genes in highly condensed regions are supposed to be turned off).
Yeast longevity genes
|
gene |
known or proposed function |
|
Lag-1 |
The Lag-1 protein product (Lag-1) regulates traffi c between the endoplasmic reticulum and Golgi complex and is required for the construction of a normal glycocalyx. The aging mechanism is unclear but may involve cell-surface signaling (mediated by the glycocalyx) that influences growth, stress resistance, and apoptosis. |
|
Ras-1 |
The Ras-1 product (Ras-1) is responsible for regulating the stress response. |
|
Ras-2 |
Its product regulates the mitochondrial retrograde response, participates in the regulation of the stress response, and is necessary for genetic stability. |
|
Rpd-3 |
Its product is a histone deacetylase that is needed for proper gene silencing and regulation. |
|
Hda-1 |
Its product is another histone deacetylase that regulates silencing of ribosomal RNA genes. |
|
Sir-2 |
Sir-2 regulates ribosomal RNA genes. |
|
Sgs-1 |
The Sgs-1 protein product (Sgs-1) codes for a DNA helicase that is required for DNA replication. This gene is homologous to the human wrn gene, which, when mutated, greatly accelerates the rate of aging. |
Note: Gene and protein naming conventions are explained in next topic.
Active regions of the genome are associated with chromatin that is acetylated; that is, the histones are modified with the addition of acetyl groups, thus marking the region as being transcriptionally active. Two yeast longevity genes, Rpd-3 and Hda-1, code for enzymes called deacetylases that remove the acetyl groups, thus converting chromatin from an active to an inactive configuration. A third gene, called Sir-2, is also responsible for gene silencing, but its mechanism of action is not clear. Damage to any of these silencing genes can shorten the life span of a yeast cell. Ribosomal RNA (rRNA) gene expression is one system that is affected by these longevity genes. Without appropriate gene silencing, production of rRNA is excessive and is not balanced by the synthesis of ribosomal proteins. The consequence is the assembly of defective ribosomes and a reduction in the efficiency of protein synthesis.
The maintenance of genetic stability, the fourth major process affected by the aging process, is provided by a host of nuclear proteins and enzymes that repair DNA damage and by many other proteins that are needed for accurate replication. One such enzyme, called a helicase, is encoded by the Sgs-1 gene. The function of a helicase is to unwind the DNA helix in preparation for replication. Mutation of this gene leads to the corruption of many genes during replication and is associated with accelerated aging.
Nematode
A nematode is a very small round worm that inhabits the soil and sometimes the digestive tracts of mammals. Mammalian parasite nematodes are known as pinworms. The nematode Caenorhabditis elegans is a popular research organism among developmental biologists and gerontologists. Several longevity genes have been identified in C. elegans, most of which are involved in an insulin-like signaling pathway. At the head of this pathway is the insulin-like receptor, encoded by the gene Daf-2.
The Daf-2 pathway mediates growth and proliferation signals necessary for the active lifestyle of an adult nematode. Mutation of Daf-2 shifts the entire physiology of the animal from active behavior to something resembling hibernation in mammals. Hibernation behavior in nematodes is known as a diapause state. Nematode diapause is characterized by a shift from active glucose metabolism (i.e., burning calories) to storage functions, such as the deposit of fat. The animal’s activity level drops, and the life span is increased by nearly 80 percent. Thus, Daf-2 is a negative regulator of life span; it is an example of the kind of gene that limits life span as a result of maximizing activity level and metabolic performance. The effects observed in Daf-2 mutants are very similar to the response of mammals to hibernation or caloric restriction. The products of other nematode longevity genes, such as Age-1, Daf-18, Akt-1, and Daf-16, transduce the signal received by the Daf-2 receptor protein (e.g., the Age-1 protein conveys the signal from the Daf-2 receptor to the interior of the cell). Consequently, a mutation in any of these genes will lead to the diapause state and extended life span.
A second pathway has been identified that affects nematode longevity. The Daf-12 gene codes for a steroid hormone receptor that is linked to a pathway that appears to regulate the stress response. Indeed, this pathway specifies resistance to heat, ultraviolet radiation, and oxidative stress. Accordingly, Daf-12 is a positive regulator of life span. A mutation in Daf-12 or in Ctl-1, a component of the pathway, shortens life span.
Fruit fly
The fruit fly Drosophila melanogaster is a popular research organism. During the 1980s researchers managed to isolate long-lived Drosophila through selective breeding. These flies showed a greater metabolic capacity and enhanced resistance to stress initiated by heat, desiccation, and ethanol vapors. In addition, they have higher activities of antioxidative enzymes, they are more efficient at utilizing nutrients, and they have enhanced stores of lipid and glycogen. Many of these features are held in common with long-lived nema-todes and yeast.
Direct support for the free radical theory of the aging process came with the isolation and characterization of Sod-1, the gene coding for superoxide dismutase. Transgenic fruit flies overexpressing Sod-1 live longer than normal and suffer much less oxidative damage induced by free radicals. Interestingly, overexpression of Sod-1 in motorneurons alone is sufficient to nearly double the mean life span of these animals. Overexpression of another gene, Mth, also increases life span. The Mth protein product, called methuselah, is a cell surface receptor that is linked to a pathway that regulates the stress response.
The retrograde response (described above) involving traffic between the cell nucleus, cytoplasm, and the mitochondria, is also involved in Drosophila aging. The Indy (I’m not dead yet) gene codes for a mitochondrial membrane protein involved in transport of Krebs cycle intermediates.
Caenorhabditis Elegans Longevity Genes
|
gene |
known or proposed function |
|
Daf-2 |
The product of this gene is an insulin-like cell membrane receptor (Daf-2). Disrupting this pathway extends life span. |
|
Age-1 / Daf-23 |
These genes code for two kinases, directly linked to the Daf-2 signaling pathway. |
|
Daf-18 |
The protein product is on the Daf-2 pathway, downstream from the Age-1/Daf-23 products. |
|
Akt-1 / Aakt-2 |
The products of these genes are on the Daf-2 pathway downstream from Daf-18. |
|
Daf-16 |
Daf-16 is a multifunction factor that is activated by the Daf-2, and Daf-12 pathways. Loss of function promotes a "hibernation" response, involving the storage of fat and glycogen that extends life span. |
|
Daf-12 |
Daf-12 is a steroid hormone receptor that is linked to a pathway important in stress resistance. A mutation in this gene shortens life span. |
|
Ctl-1 |
The protein product is a cytoplasmic enzyme (catalase) on the Daf-12 stress-resistance pathway. |
Note: Gene and protein naming conventions are explained in next topic.
Drosophila Longevity Genes
|
GENE |
KNOWN OR PROPOSED FUNCTION |
|
Indy |
This gene codes for a mitochondrial membrane protein involved in transport of Krebs cycle intermediates. The loss of function increases life span by reducing the availability of nutrients (caloric restriction). |
|
Sod-1 |
The protein product is superoxide dismutase (Sod). Overexpression increases life span by enhanced inactivation of free radicals. |
|
Mth |
Its product codes for a cell membrane receptor called methuselah, which enhances the stress response, thus increasing life span. |
|
Chico |
The protein product, Chico, is a hormone similar to mammalian insulin. Loss of function increases life span through caloric restriction. |
|
Inr |
This gene codes for the Chico receptor. Lose of function has the same effect as a Chico mutation. This receptor is very similar to the nematode DAF-2 receptor. |
|
Sugar baby |
The protein product is a maltose permease. Overexpression increases life span by shifting metabolism away from glucose, thus invoking partial caloric restriction. |
Note: Gene and protein naming conventions are explained in next topic.
A mutation in the Indy gene blocks import of these compounds, with an effect similar to caloric restriction—a near doubling of life span. Insulin and insulin receptors modulate life span in Drosophila much as they do in nematodes. The Drosophila genes Chico and Inr (Insulin receptor) encode an insulin protein and insulin receptor that are very similar to those found in nematodes and mammals. Mutations in Chico or Inr have the same physiological effects as described for the Daf-2 gene in nematodes. The Sugar baby gene achieves a similar though muted effect on life span. This gene codes for a maltose permease, an enzyme that enhances the uptake of maltose into cells. Overexpression of this gene shifts the animal’s physiology away from glucose utilization, thus mimicking the effects of caloric restriction. In this case, the increase in life span is about 20 percent, compared with the more than 80 percent increase observed in Inr mutants.
Mouse
The most consistent way to extend the life span of a mammal is by caloric restriction. Such experiments have extended the life spans of mice and rats by up to 50 percent. Moreover, these calorie-restricted animals show similar metabolic responses observed in yeast, nematodes, and fruit flies, including resistance to stress. In addition, calorie-restricted rodents show a postponement of age-related diseases, such as cancer, and have an increased lifetime metabolic capacity. These changes, like the hibernation response in nematodes and flies, are due to more efficient utilization of glucose and a shift toward deposit of fat and glycogen.
Three mouse genes have been identified that, when mutated, extend life span in a manner similar to caloric restriction. The gene Prop-1 ("Prophet of pit-1") codes for a protein that regulates another gene, Pit-1, that codes for a pituitary-specific transcription factor. Mutation of Prop-1 or Pit-1 leads to developmental arrest of the pituitary gland, thus drastically reducing the normal levels of growth-inducing hormones such as growth hormone (GH) and thyroid hormone (TH). In the absence of these hormones, cells cannot utilize glucose or amino acids to promote growth and maturation. Consequently, Prop-1 mutants are dwarfs, but they have an extended life span. This mutation mimics a calorie-restricted diet that begins in the womb.
Lab mouse
A second type of longevity gene has been identified in mice. This is the P66shc gene, which codes for a component of a signaling pathway that regulates the stress response and apoptosis. As with the other positive longevity genes already described, overexpres-sion of this gene increases life span, while animals possessing a normally expressed P66shc have shorter life spans.
Human
Identification of longevity genes in lower organisms has stimulated a search for similar genes in the human genome. The human homolog of yeast Lag-1 has already been cloned and is located on chromosome 19. Although the sequence homology is low, it can replace the yeast gene where it performs a longevity function. Consequently, human Lag-1 may be thought of as a human longevity gene, although much work is needed to confirm its function in humans.
Mouse longevity genes
|
GENE |
KNOWN OR PROPOSED FUNCTION |
|
Prop-1 |
The protein product is a regulator of a pituitary-specific transcription factor (Pit-1). Inactivation leads to poor development of the pituitary and production of pituitary hormones, particularly growth hormone. Mutated Prop-1 increases life span by about 50 percent. |
|
Pit-1 |
This gene codes for Pit-1, a protein transcription factor. The inactivation of Pit-1 has the same effect as a Prop-1 mutation. |
|
P66shc |
The protein product is a component of a signal transduction pathway that makes cells resistant to apoptosis and oxidative stress. |
Note: Gene and protein naming conventions are explained in next topic.
Perhaps the most striking similarity between longevity genes in humans and lower organisms is the yeast Sgs-1 gene and the human Wrn gene. The Sgs-1 gene codes for a helicase, and when mutated, can accelerate the aging process. Werner’s syndrome is a disease in humans that is also associated with accelerated aging. The gene responsible for this disease, called Wrn (for Werner’s syndrome), has been identified. The protein product of the Wrn gene is a helicase, not the same helicase encoded by the Sgs-1 gene, but a member of the same family, possessing a similar function. Mutations in these two genes provide dramatic evidence in support of the connection between life span and the maintenance of genetic stability.
Summary
The search for longevity genes has identified four processes that influence life span. They are metabolic control, resistance to stress, gene disregulation, and genetic stability. Evidence supporting the involvement of metabolic control comes from the roles of Lag-1 in yeast, Daf-2 in nematodes, Indy and Sod-1 in Drosophila, and Prop-1 in mice. Resistance to stress is a function of several longevity genes, such as Ras-2, Daf-12, Mth, and P66shc. Gene disregulation, as a mechanism of aging, has been clearly demonstrated in yeast with the isolation of three histone deacetylase genes, Rpd-3, Hda-1, and Sir-2. Finally, the relationship between genetic stability and life span is indicated by the effects of Sgs-1 mutants in yeast and the human disease known as Werner’s syndrome, which is associated with accelerated aging and is caused by the gene Wrn, a homolog of Sgs-1.
This collection of genes, small though it is, has given a powerful boost to aging research and provides an important conceptual framework that future research may follow. The goal is to isolate even more longevity genes from lower animals, and then to find their counterparts in the human genome. This work has already begun with the isolation of human Lag-1. The characterization of all longevity genes will improve our understanding of cellular senescence. Manipulation of these genes might also provide a way to reverse some of the effects of the aging process.