Information Technology Reference
In-Depth Information
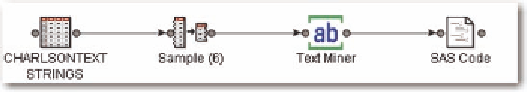
Figure 5. SAS text miner
Figure 6. Scoring all data
dx8,dx9,dx10,dx11,dx12,dx13,dx14,dx15);
Procedures=catx(' ',pr1,pr2,pr3,pr4,pr5,pr6,pr7,
pr8,pr9,pr10,pr11,pr12,pr13,pr14,pr15);
Run;
Then, we use the following diagram in SAS Enterprise Miner (Figure 5).
We generally use a sample of the data because of the amount of computer resources required to per-
form Text Miner. We can then score the remaining data, meaning that we use a small sample to define the
clusters and then we use the remaining data as test data (Figure 6). The metadata node is used to change
the definition of the cluster value as a target variable; the MBR node is used to define the prediction of
the cluster. The score node is used to define the cluster value for the remaining data.
In order to run the process shown in Figure 6, the data role for the scored data must be changed to
“score”. The Text Miner node has some defaults that need to be changed in order to work with ICD9
codes (Figure 7).
In particular, we must allow Text Miner to cluster using numbers, since numbers are all that are
available. Generally, the default is not to use numbers. We also don't want Text Miner to attempt to dis-
tinguish between different parts of speech since all of the ICD9 codes are nouns, and we don't want to
use noun groups. Once the singular value decomposition is completed, we can go ahead and cluster the
patient records. For a patient severity index, we want to define a small number of clusters, so we define
the total number of clusters exactly. In this case, we use a total of ten diagnosis clusters.
We can similarly cluster based upon procedures, although we have to make a slight modification in
the dataset, or we will overwrite the cluster numbers. We use the created dataset, emws.text_documents
and define a new dataset with the code:
Data nis.diagnosisclusters (drop=_SVD_1-_SVD_100 prob1-prob100);
Set emws.text_documents;

Search WWH ::

Custom Search