Agriculture Reference
In-Depth Information
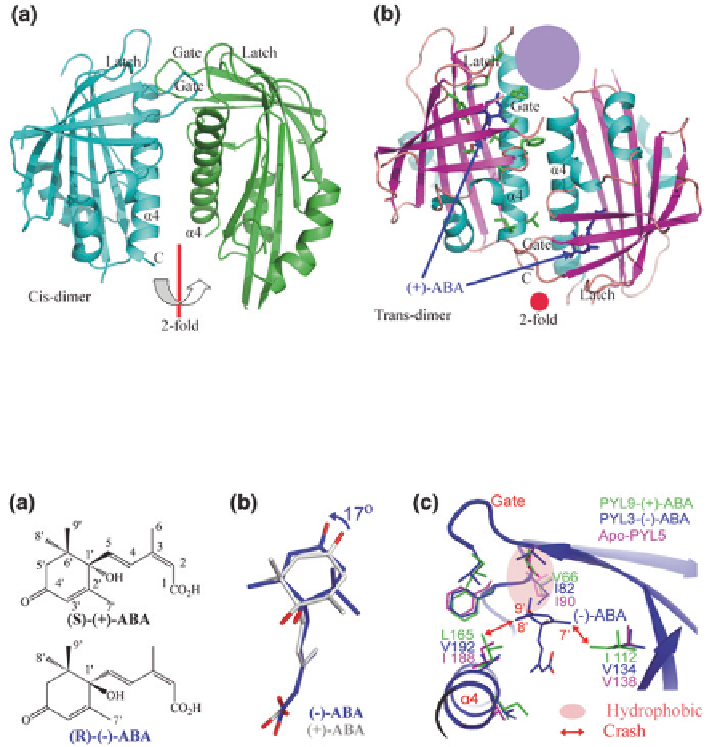
Fig. 7.4
Formation of the PYL3 trans-homodimer with ligands.
a
Apo-PYL3 is a cis-homodi-
mer, two protomers are related by a twofold rotation axis, parallel to the plane of the page.
b
The PYL3-(
+
)-ABA complex structure is a trans-homodimer. The (
+
)-ABA (
color blue
) bind-
ing pocket in ligand-bound PYL3 is exposed to the solvent and cycled (
purple
). Two protomers
are related by a twofold rotation axis, perpendicular to the plane of the page (Zhang et al.
2012
)
Fig. 7.5
Structure alignment of (-)-ABA with (
+
)-ABA.
a
Chemical structure of (
+
)-ABA and
(-)-ABA.
b
Superposition of (
+
)-ABA and (-)-ABA from their complex structures in PYL3.
There were partially rotation and shift between the rings in both ABA.
c
Superposition of apo-
PYL5, PYL3-(-)-ABA and PYL9-(
+
)-ABA indicated that the major variant residues underlain
the favor of PYL binding with (-)-ABA. Two bulk side chains of I112 and L165 in PYL9 seri-
ously collided to 7′ and 8′ methyl groups of (-)-ABA, respectively. The stereo constraints were
vanished in PYL5 because of two corresponding small side chains. On the other hand, V66I in
PYL9 would give a strong coordination with 8′ and 9′ methyl groups in (-)-ABA through a strong
hydrophobic network (Zhang et al.
2013
)
PYL9-(
+
)-ABA were determined to uncover the reason (Zhang et al.
2013
). By
superimposing these three structures, we found that the binding orientation and
pocket of (-)-ABA in PYLs are obviously different from those of (
+
)-ABA (see
Fig.
7.5
b), which might deny the “flip” hypothesis. Structural and biochemical
investigations showed the major variable residues surrounding the mono-methyl
and di-methyl groups of ABA cyclohexene ring might underlay the preference of
Search WWH ::

Custom Search