Agriculture Reference
In-Depth Information
to date. These data confirmed that the PYLs family belongs to a branch of the Bet
v I superfamily, which possesses a large and conserved hydrophobic ligand-bind-
ing pocket.
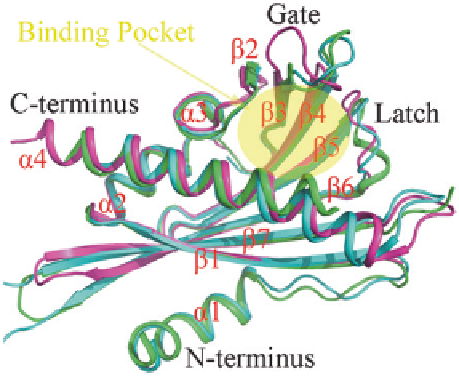
From the published apo-PYLs structures, we know that all the PYLs share a
highly similar helix-grip structure, in which the
ʱ
2 helix and the C-terminal
ʱ
4
helix enclose a seven-stranded
ʲ
-sheet to create a ligand-binding pocket, which
accommodate the solvent (Fig.
7.1
). Two highly conserved loops encompass
the ligand-binding pockets: the residues SGLPA 'gate' loop (also refers to CL2
loop) between the
ʲ
3 and
ʲ
4 strands; the residues HRL 'latch' loop (also refers
to CL3 loop) (Melcher et al.
2009
; Yin et al.
2009
) between the
ʲ
5 and
ʲ
6 strands
(Fig.
7.1
). These two loops are very important for ABA binding.
Usually, the conformation of the receptor molecule changes when binding to
ligand. This alters the interaction of the receptor molecules with associated bio-
chemicals and, in turn, leads to a cellular response mediated by the associated
biochemical pathway. From the ABA-bound structures of PYR1, PYL1, PYL2,
PYL3, PYL9, PYL10 (Miyazono et al.
2009
; Santiago et al.
2009
; Yin et al.
2009
;
Hao et al.
2011
; Zhang et al.
2012
,
2013
; Li et al.
2013
), the ABA is encaged in
the binding pocket by a combination of hydrophobic interactions and water-medi-
ated hydrogen bonds (see Fig.
7.2
b, c). First, the acidic head group of ABA faces
innermost, forms a conserved water-mediated hydrogen bond network with polar
residues of PYLs, defining the location of the pentadienoic acid moiety and the
hydroxyl group of ABA in the binding pocket, and forming charge interactions
with a conserved lysine (such as K59 in PYR1, K86 in PYL1, K79 in PYL3) (see
Fig.
7.3
). These polar interactions help to locate the ABA molecule. However, this
lysine residue is replaced by a glutamine in PYL13, consistent with the lack of
evidence for binding ABA. In the recently published structure of PYL13 (Li et al.
2013
), we know that in addition to the lost lysine residue, there would be a steric
clash between the aromatic ring of Phe71 residue in PYL13 and the hydrophobic
moiety of ABA. An invariant residue leucine is located in the corresponding site
Fig. 7.1
Structure
comparison of PYR1, PYL3
and PYL10. PYR1 (pdb
ID: 3K90), PYL3 (pdb ID:
3KLX) and PYL10 (pdb
ID: 3UQH) are shown in
green
,
cyan
and
magenta
,
respectively. Binding pocket
is shown in
yellow ellipse
Search WWH ::

Custom Search