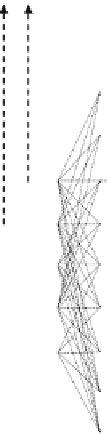Biology Reference
In-Depth Information
G
‡
D
G
ij
‡
D
G
j
‡
D
G
i
‡
1
1
2
2
G
3
3
4
4
5
5
G
i
G
j
G
ij
T
x
D
x
(T
x
D
x
)(T
Y
D
Y
)
T
Y
D
Y
Fig. 12.31 A diagrammatic representation of the Gibbs free energy levels of T
X
D
X
and T
Y
D
Y
complexes (see G
i
and G
j
) and their higher-order complex (T
X
D
X
)(T
Y
D
Y
) enzyme complexes (see
G
ij
). The Gibbs free energy level of the common excited state is denoted by G
{
.
D
G
i
is the Gibbs
free energy difference between the excited and ground states of the ith enzyme system, i.e.,
D
G
i
¼
G
{
-G
i
. It is assumed that T
X
D
X
and T
Y
D
Y
complexes have five energy levels and the
(T
X
D
X
)(T
Y
D
Y
) complex has nine free energy levels (see Fig.
12.29
for related discussions).
T
¼
tra
nscriptosome,
D
¼ degradosome
, and X and Y are two different RNA trajectories
(or ribons) (see [mRNA]
X
and [mRNA]
Y
in Fig.
12.27
) signal the underlying
control mechanism
that is thermally activated and functions during the observa-
tional period of 850 min (see Step 9 in Fig.
12.27
). Therefore, it seems logical to
invoke at least two levels of metabolic control in budding yeast - the first-order
control catalyzed by the individual (TD) complexes and the second-order control
exerted by the pathway-wide (T
X
D
X
)(T
Y
D
Y
) complexes. By “pathway-wide,”
I mean the system of transcriptosomes and degradosomes belonging to a given
metabolic pathway. Since each metabolic pathway usually involves 10-30
enzymes, the number of all possible pairs of enzymes, X and Y, that belong to
a pathway would range from about 50 to 500 (based on the equation n(n
1)/2,
where n is the number of different enzymes belonging to a pathway). If all these
possible pairs of enzymes function coherently, as evidenced by the correlations
found among the intra-pathway mRNA trajectory pairs shown in Table
12.12
,