Biology Reference
In-Depth Information
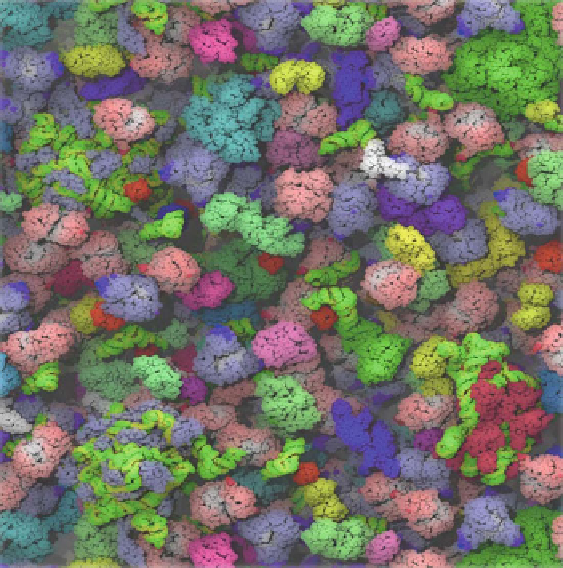
Fig. 12.28 The computer model of the cytoplasm of
E. coli
that contains over 80% of the 50
different types of the most abundant proteins in the organism (McGuffee and Elcock 2010).
RNA molecular are colored
green
and
yellow
(Reproduced by permission of Adrian Elcock
)
(26,000 Da), for example, is only five, at any instance, while, for the 50 S ribosomal
subunit (1,355 KDa), it is more than 25. It is clear that the Gibbs free energy levels
of enzymes will critically depend on their microenvironment inside the cell.
The kinetic trajectories of mRNA molecules of budding yeast undergoing
glucose-galactose shift are not random but exhibit regularities as amply
demonstrated by Figs.
9.1
,
12.2a
and
12.3
, especially the mRNA trajectories
shown in the last two figures that are function-related. This indicates that the
transcriptosome
(T) and the
degradosome
(D) responsible for the nonrandom
behavior of an mRNA trajectory must be
coupled
(see Steps 7 and 8 in
Fig.
12.27
, where the wiggly lines indicate the functional coupling between T and
D), although the molecular mechanisms underlying such a coupling are not obvi-
ous. If these two enzyme systems are not coupled, the associated mRNA trajectory
is expected to behave randomly and unpredictably, contrary to our observations.
Since the rate constant of an enzyme is the exponential function of its ground-state
Gibbs free energy level (see the right panel of Fig.
11.28
andEq.
12.42
), itwould follow
that the rate of the synthesis of an mRNAmolecule depends on the ground-state Gibbs
free energy level of the associated transcriptosome (see Steps 1 and 2 in Fig.
12.27
),