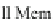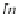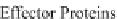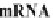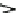Biology Reference
In-Depth Information
Fig. 11.10 The cell viewed as a system (or network) of
molecular machines
that has evolved to
carry out two main functions: (1)
signal transduction
(see Steps 1, 2, and 3) to retrieve select
“atomic survival programs” (ASPs) stored in DNA,and (2)
gene expression
(see Steps 4, 6, and 8 )
to transduce ASPs into survival actions or mechanisms, otherwise known as cell functions. See
the cell to be able to make such complex decisions, it would need similarly
complex internal states, according to the Law of Requisite Variety (see Sect.
machinery carrying out the required selective actions. So the well-recognized
complexities of signal transduction pathways can now be understood as resulting
from the cell's need to retrieve select ASPs from the nucleus and to execute them
into actions in order to survive under very complex environmental conditions that
are constantly changing. We may represent this idea schematically as shown in
Fig.
11.10
:
Step 1 is where the external signal is recognized through the receptors embedded
in the cell membrane or nuclear receptors present in the cytosol and the nucleus (not
shown), and in Step 2 the original signal is transformed to retrieve appropriate
information from DNA in the nucleus in Step 3. In Step 4, selected ASPs are
retrieved from chromosomes and transcribed into mRNA expressed at right times
and for right durations. It is important to note here that mRNA levels are functions of
not only the rates of transcription (Step 4) but also of the rates of transcript degrada-
tion (Step 5). Ignoring this simple fact has led to many false positive and false
negative conclusions in the field of microarray technology as pointed out in Ji et al.
(2009a). Similarly, effector protein levels are functions of both translation (Step 6)
and protein degradation (Step 7). Step 8 represents catalysis, the all-important step
where chemical reactions are catalyzed to generate free energy that drives all
intracellular molecular processes. Abbreviations are as follows: STP
¼
signal
transducing proteins; IDSs
intracellular dissipative structures such as intra-
cellular ion gradients (Chap.
9
) and mechanical stress gradients of cytoskeletons
(Ingber 1998). Effector proteins include transcription initiation/elongation
complexes (acting on Step 4 thus constituting a feedback loop), molecular
motors and pumps, kinases, phosphatases, synthetic enzymes, cytoskeletons, etc.
¼