Biology Reference
In-Depth Information
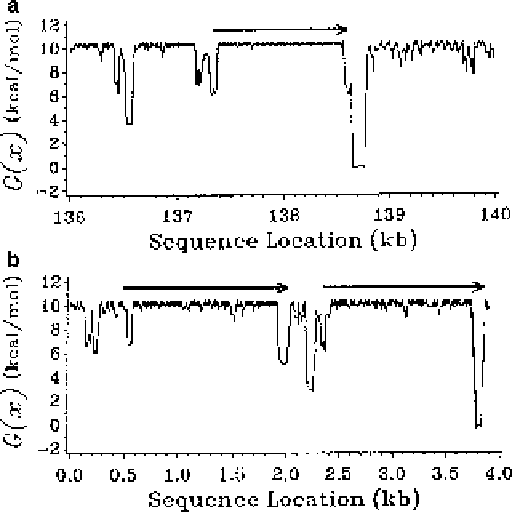
Fig. 8.3
Mechanical strains of DNA localized at sequence-specific sites within circular DNA
duplexes. The
x
-axis records the nucleotide positions along the DNA duplex and the
y
-axis records
the Gibbs free energy required to separate the based pairs located at position
x
along the DNA
duplex chain. Notice that the base pairs located near the 3
0
-end (i.e., the right-hand end of the
arrow
) of some genes are already completely separated (see position 138.7 in (
a
) and 3.56 in (
b
))
binding-induced gene expression. Similar ideas have been proposed by others
(Volkov 1996; Hisakado 1997; Cuevas et al. 2004; Alvarez et al. 2006). The TFCC
hypothesis provides a rational explanation for the well-known phenomenon
that a circular DNA duplex must exist in a supercoiled state before its genes can
be transcribed or replicated (Benham 1996a, b).
In the early 1990s, C. Benham developed a statistical mechanical equation to
describe the dynamics of the mechanical strains introduced in circular DNA
duplexes (Benham 1996a, b; Benham and Bi 2004). His computational results
indicated that the so-called stress-induced duplex destabilizations (SIDDs) (equiv-
alent to
a <
0) were not randomly distributed along the circular DNA duplex but
were localized mainly to the 5
0
and 3
0
ends of RNA coding regions. Three examples
of SIDDs are shown in Fig.
8.3
(see the directed arrows), where the downward
deflections indicate the decrease in the Gibbs free energy needed for strand separa-
tion due to the localized destabilization induced by mechanical strains. Thus, both
the
sequence-specificity
and the
mechanical energy
stored in DNA make SIDDs
excellent examples of the more general notion of
conformons
invoked two decades
earlier and restated in Statement 8.7 (Green and Ji 1972a, b; Ji 1974b, 2000).
A more direct experimental evidence for the production of conformons from
ATP hydrolysis was recently reported by Uchihashi et al. (2011; Junge and M
€
uller