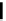Biology Reference
In-Depth Information
STRUCTUR
A
L BENCHMARK
3D Structures
Ref
erence
MSA
Select Protein
Family
Structure
DB
Reference-free structural benchmark
Ref
erence-
ba
sed stru
ctu
ral benchm
ark
SIMULATION
Compare
Implied by Inferred MSAs
3D Structure Overlap
Compare Inferred MSAs
with
Reference MSA
Select Simulation
Parameters
CONSISTENCY TEST
Real Data
Synthetic Data
Select Protein
Family
Sequence
DB
Simulator
True MS
A
Clustal
Prank
Ma t
etc.
...
Evaluate
Consistency
of Inferred MSAs
PHYLOGENETIC TESTS
Species Tree Discordance
T
es
t
Compare Inferred MSAs
with
True MSA
Minimum Duplication Test
Select Protein
Family
Select Reference
Species Topology
Select Group of
Orthologous Proteins
Compare Inferred Trees
with
Count Min
Number of Duplications
Implied by Inferred Trees
Reference Topology
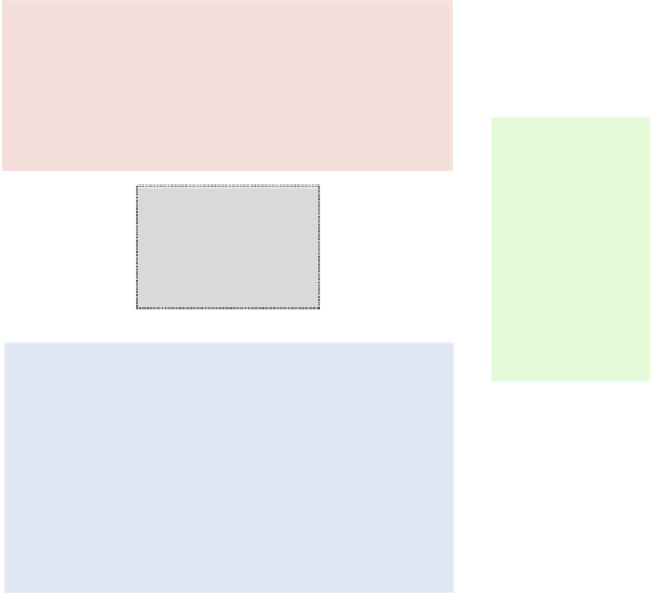
Fig. 1 Schematic of the four main MSA benchmarking strategies of this review: for each approach, the
benchmarking process starts from the corresponding downward-pointing arrow and involves alignment by
different MSA methods (gray box in center, illustrating example aligners that may be benchmarked)
l
Solvable
, in that it provides sufficient challenge to differentiate
between poor and good performances, while remaining a tracta-
ble problem.
l
Scalable
, so that it can grow with the development of MSA
programs and sequencing technologies.
l
Accessible
, in order to be widely used by developers and users.
l
Independent
from the methods used by programs under test,
as benchmark datasets should avoid any overlap with the heur-
istics chosen for construction of MSA in order to constitute an
objective reference.
l
Evolving
, to reduce the possibility of developers adapting
their programs to a particular test set over time, thus artificially
inflating their scores.
Although MSA methods employ different computational solu-
tions to reconstruct sequence alignments, their performance needs
to be assessed on the same benchmarks in order to be objectively
evaluated and compared. In this chapter, we consider four broad
MSA benchmarking strategies (Fig.
1
):
1. Benchmarks based on simulated evolution of biological
sequences, to create examples with known homology.