Biology Reference
In-Depth Information
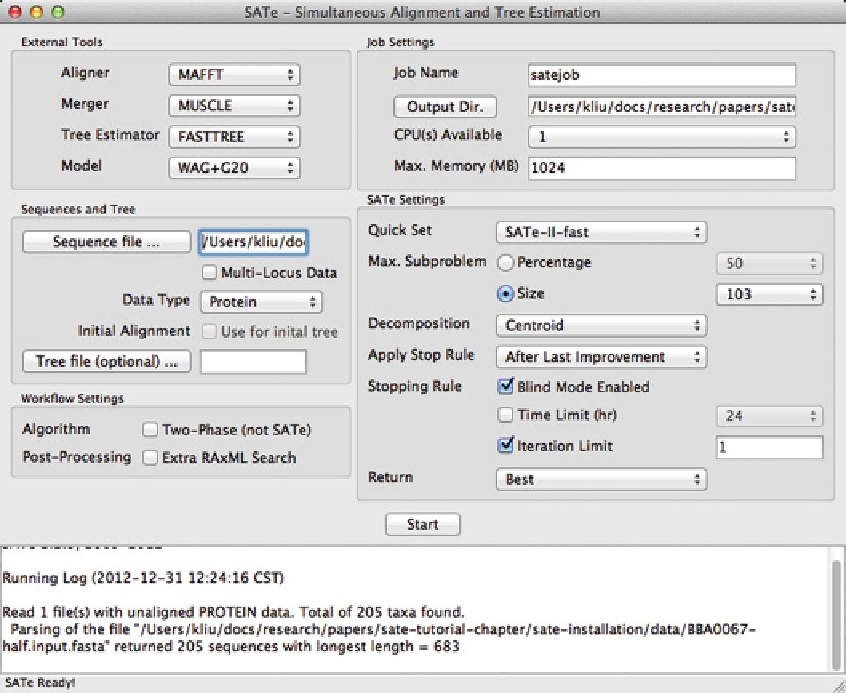
Fig. 5 The start of a SAT ´ analysis of an amino acid dataset
1. Very large datasets with tens of thousands of sequences or more
pose a special computational challenge. Changing software
settings is recommended in this instance, although the optimal
settings for a particular dataset depend upon many factors.
Thus, while we provide specific suggestions for this case,
experimenting with software settings is also advisable, with
the caveats described in
Notes 5
through
7
. See the discussion
above (for “large dataset analyses”) for some explanations for
why we make the following recommendations.
2. If an alignment and tree are already available, we recommend
providing them to SAT´. This recommendation is strongly
recommended for very large datasets (with 10,000 sequences
or more), but beneficial for all analyses.
3. Otherwise, we recommend computing an initial alignment
using either MAFFT's PartTree algorithm or Clustal Omega;
these tools are not available within the GUI usage of SAT
´
, and
so this will need to be done offline. For an input file named
8.7 Advanced
Analysis: Very Large
Datasets (More Than
10,000 Sequences)