Biology Reference
In-Depth Information
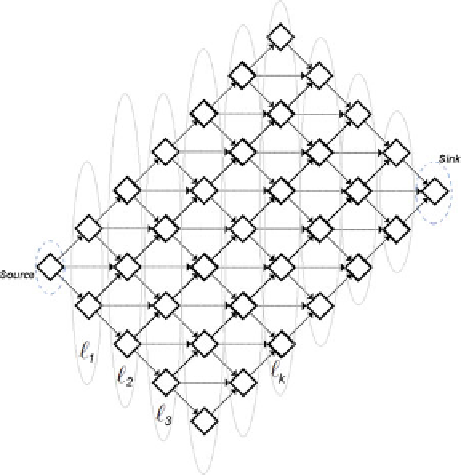
Fig. 4 Topological equivalence of multilayer networks and alignment graphs
Using the bottom-up recursion methodology of dynamic
programming, we can start with calculating the score from the
source to its children (with zero initials), and save these paths and
their scores to be used in the next iterations. The computation
terminates when the sink layer is reached and the optimal path is
found. This dynamic programming algorithm lets the computer to
deal with only the path combinations of adjacent nodes in each
iteration instead of computing the scores of every possible path.
Clearly we can see that addition of layers increases the number of
computations nearly in linear fashion given a constant average
connectivity, whereas the search space increases in exponential
fashion. This is the implication of the reduction in computational
complexity.
The graphical representation of pairwise sequence alignment
has the same multilayered network structure defined above
(Fig.
4
). We can see the topological equivalence by assigning the
top-left cell as the source, bottom-right cell as the sink, and the cells
f
k
.
A property has to hold in order to satisfy the equality of optimal
path finding and sequence alignment problems. This is the additive
property of the path scores. In terms of sequence alignment, each
pairing in the alignment (the scores of individual edges) has to be
independent of each other, and the total alignment score is
obtained by summing the up scores of each pair. Fortunately,
such a scoring scheme is biologically plausible and the currently
preferred alignment score calculation method. An alignment is
accepted to be corresponding to a parsimonious molecular
c
ð
i
;
j
Þ :
i
þ
j
¼
k
g
as the middle layer
'