Biology Reference
In-Depth Information
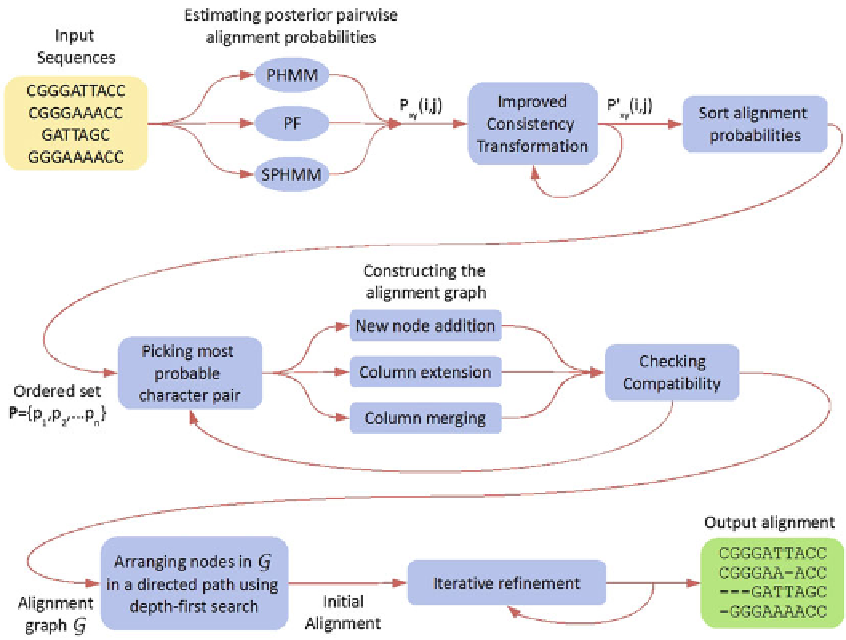
Fig. 1 Diagrammatic overview of the alignment steps in PicXAA
progressive alignment techniques suffer from. In the following,
we give a brief summary of the alignment steps that are involved
in PicXAA. Fig.
1
provides a diagrammatic overview of the align-
ment process of PicXAA.
Suppose we have estimated the posterior pairwise alignment
probabilities for all residue pairs in every possible pair of sequences
x
;
y
in a given sequence set
S
. We denote this probability as
Px
i
2.1 Improved
Probabilistic
Consistency
Transformation
, where
x
i
2 x
a
jx
;
y
y
j
2
is a residue in sequence
x
,
a
means that the
residues
x
i
and
y
j
are aligned in the true (unknown) alignment
a
.
These probabilities can be computed using various approaches,
such as pair-HMMs (hidden Markov model) [
10
], partition func-
tion based methods (
see Note 8
for parameters used in this scheme)
[
21
], and structural pair-HMMs [
15
](
see Notes 1
-
3
,
6
and
7
for
more details on these methods). Given these pairwise alignment
probabilities
Px
i
y
j
2 y
is a residue in sequence
y
, and
x
i
y
j
2
a
jx
;
y
, PicXAA updates them using an
improved probabilistic consistency transformation. The probabilis-
tic consistency transformation (PCT) attempts to enhance the
reliability of the estimated pairwise residue alignment probabilities
y
j
2