Biology Reference
In-Depth Information
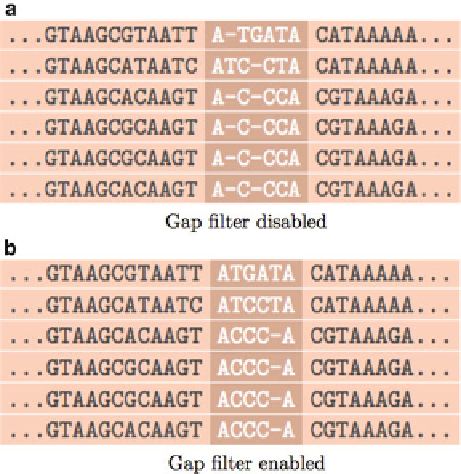
Fig. 7 Effect of post-alignment processing with the gap filter. Sometimes
non-homologous regions appear with an interleaving of gap-abundant
columns. This is due to a combination of alignment order and pairwise scoring
parameter selection. Many times the non-homologous regions are not of much
interest, in which case this doesn't matter. However, we have provided a post-
processing step that scans the MSA in search for columns composed of gaps and
with neighboring columns also with gaps, but in different rows. When found, the
gaps are blindly shift in order to compress these non-homologous regions
together
After the MSA has been created,
GRAMALIGN
provides a post-
processing step designed to perform a blind shift of gaps. The
goal is to reduce non-homologous regions containing interleaved
columns composed largely of gaps. For example
see
Fig.
7
, which
depicts an original alignment in (a) and the result of enabling the
filter in (b). The locally-affected region is highlighted. By default
this post-processing gap adjustment step is disabled based on the
default values of the following options.
Option -t <value>
3.8 Alignment Gap
Filter Options
: Specify the percentage of gaps in a column
before a blind adjustment can occur. At the end of the MSA
algorithm, the alignment is scanned for columns containing at
least as many gaps as specified via this percentage (e.g., 0
¼
0%
¼
zero gaps in the column, 1.0
¼
100 %
¼
column with all gaps,
0.5
at least half of the column entries are gaps).
If any column contains at least this many gaps, a surrounding
window (specified in the -w option) of columns is checked for
possible gaps that may be shifted into the current column. To
disable this action, simply set this value to 1.0, thus setting the
threshold to be columns that contain nothing but gaps, which are
¼
50 %
¼