Biology Reference
In-Depth Information
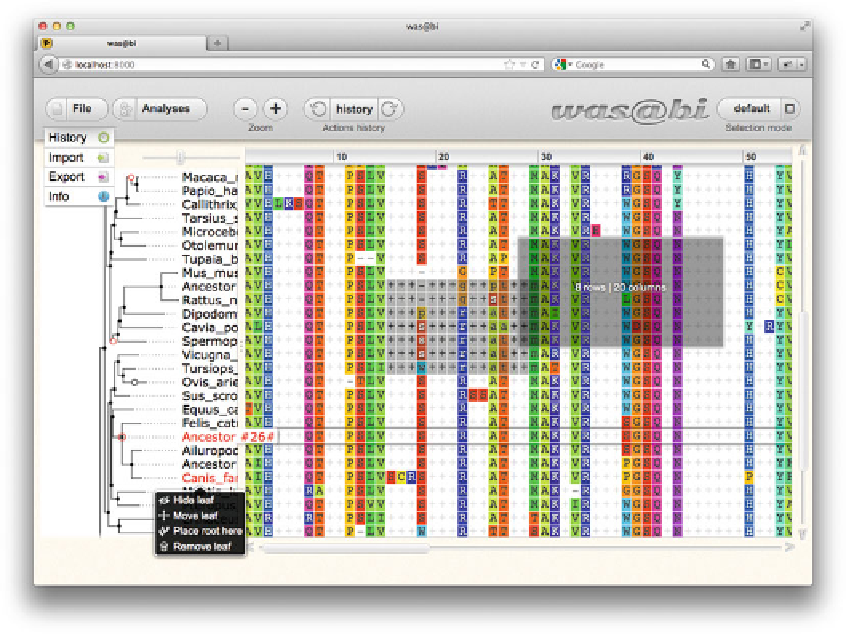
Fig. 7 Wasabi provides a web browser-based graphical interface to PRANK and related evolutionary sequence
analysis programs. The visualization and analysis of multiple sequence alignments should be performed in the
context of the phylogeny relating the sequences. In addition to integrated alignment and analysis with
PRANK
and related tools, the Wasabi browser interface allows for manipulating the sequence phylogeny, showing and
hiding the inferred ancestral sequences and both automated and manual masking of parts of the alignment.
Wasabi can be found from the
PRANK
home page at http://code.google.com/p/prank-msa/
and provides an easy-to-use access to
PRANK
and related methods
(Fig.
7
). This tool and all other methods discussed here can be
References
1. Sankoff D (1975) Minimal mutation trees of
sequences. SIAM J Appl Math 28:35-42
2. Ogurtsov A, Sunyaev S, Kondrashov A (2004)
Indel-based evolutionary distance and mouse-
human divergence. Genome Res 14:1610-1616
3. Hogeweg P, Hesper B (1984) The alignment
of sets of sequences and the construction of
phyletic trees: an integrated method. J Mol
Evol 20:175-186
4. Feng D, Doolittle R (1987) Progressive
sequence alignment as a prerequisite to correct
phylogenetic trees. J Mol Evol 25:351-360
5. L¨ytynoja A, Goldman N (2008) Phylogeny-
aware gap placement prevents errors in
sequence alignment and evolutionary analysis.
Science 320:1632-1635
6. L¨ytynoja A, Goldman N (2005) An algorithm
for
progressive multiple
alignment
of