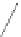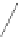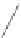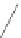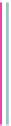Biology Reference
In-Depth Information
Dense vs. sparse sampling:
TCA
G
- TCG
A
TCAGTCG
A
G
A
TCAG-TCG
B
TCA- -TCA
C
TCA--TCG
D
CCA- - -CG
E
CCA-CTCG
TCATC
G
1
TCA - - TC
A
B
TCATCA
Z
B
G
A
T
CATCG
TCA - - TCG
2
C
TCATCG
Y
Y
C
C
T
C
CATCG
CCA - - - CG
3
D
CCACG
T
X
D
CCATCG
CCA -
C
TCG
E
4
CCACTCG
W
E
C
1
2
Z
3
Y
3
X
A
TCAGTCG
A
C
G
T
A
G
T
A
C
G
T
A
C
G
T
A
G
T
A
C
G
T
A
C
G
T
A
G
T
-
A
C
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
G
T
A
G
T
A
G
T
A
G
T
A
G
T
B
TCA-TCA
C
TCA-TCG
D
CCA - - CG
E
CCA - CTCG
Z
A
C
G
T
A
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
G
T
A
G
T
A
C
T
A
C
T
A
C
T
Y
G
A
C
T
C
G
T
A
C
T
C
G
T
X
A
A
C
T
C
G
T
A
C
T
C
G
T
W
A
C
T
A
C
T
C
G
T
A
C
T
A
C
T
C
G
T
T
C
G
T
C
G
T
C
G
T
C
G
T
TCA
G
-TCG
A
TCAGTCG
A
A
TCAGTCG
B
G
TCA-TCA
D
CCA- -CG
E
T
CATC
G
1
TCA - - TC
A
B
TCATCA
Z
C
G
A
C
T
C
CATCG
CCA - - - CG
2
CCACTCG
D
CCACG
X
X
D
T
CCATCG
CCA -
C
TCG
E
3
CCACTCG
C
W
E
1
A
TCAGTCG
2
Z
3
X
A
A
C
T
A
C
T
A
C
T
A
C
T
A
C
G
T
A
C
T
A
C
T
C
G
T
C
G
T
C
G
T
C
G
T
C
G
T
C
G
T
C
G
T
B
TCA-TCA
D
CCA - - CG
E
CCACTCG
A
C
T
Z
A
C
T
A
C
T
A
C
G
T
C
G
T
C
G
T
C
G
T
X
A
C
G
T
A
G
T
A
C
G
T
W
A
C
G
T
A
C
G
T
A
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
G
T
A
C
G
T
A
G
T
A
G
T
Fig. 6 Correct identification of independent insertion and deletion events requires closely-related sequences.
With a dense sampling of sequences (top) each insertion and deletion event can be identified using the
outgroup information from the next alignment and the correct homology is recovered. With a sparser sampling
(bottom), the insertion in A cannot be identified because of a deletion at an adjacent position in D. As a result,
the independent insertions in A and E are incorrectly matched
Similar heuristics unfortunately cannot be provided for missing
data in other parts of the sequences.
The phylogeny-aware alignment algorithm assumes that each
alignment gap is caused by one insertion or deletion event and that
the very next alignment provides information to distinguish
between the two types of events. When the sequences are relatively
closely related (and, as stated previously, the alignment order is
correct), these assumptions are typically valid. If the sequences are
more diverged, the chances of independent insertion and deletions
events at near-by positions in the adjacent evolutionary branches
become significant. As a result of this, either the gap created in the
first alignment may be a combination of two or more separate
events, or the subsequent alignment of an outgroup sequence
fails to confirm the event as an insertion or a deletion due to an
overlapping independent event in the neighboring branch (Fig.
6
).