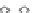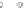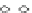Biology Reference
In-Depth Information
Wrong alignment order:
Right order
Wrong order
TCA - TCG
A
TCATCG
A
TCATCG
A
TCATCG
1
1
TCA - TCG
TCATCG
CCAGTCG
B
Z
B
Z
C
G
T
CA
G
TCG
C
CAGTCG
2
2
C
CCAGTCG
TCATCG
Y
C
Y
B
T
C
TCAGTC
G
TCAGTC
A
3
3
D
TCAGTCA
TCAGTCA
X
D
X
D
G
A
1
2
3
A
TCATCG
A
G
T
A
G
T
Z
A
C
G
T
-
A
C
G
T
Y
A
C
G
T
A
C
G
T
C
G
T
C
G
T
C
G
T
A
C
G
T
C
G
T
C
G
T
C
G
T
A
TCA-TCG
B
TCA-TCG
C
CCAGTCG
D
TCAGTCA
B
TCATCG
B
CCAGTCG
D
TCAGTCA
A
C
G
T
Z
A
C
G
T
A
C
G
T
C
G
T
C
G
T
C
G
T
Y
A
C
T
A
C
T
A
C
T
X
A
C
T
A
C
T
A
C
T
A
C
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
G
T
A
G
T
A
G
T
1
2
Z
3
Y
A
TCA-TCG
A
C
T
A
C
T
A
C
T
A
C
G
T
A
C
T
A
C
G
T
A
C
T
A
C
G
T
-
A
C
T
A
C
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
C
G
T
A
TCA- -TCG
C
CCAG-TCG
B
TCA- -TCG
D
TCA-GTCA
C
CCAGTCG
B
TCA-TCG
D
TCA-GTCA
Z
A
C
T
A
C
G
T
A
C
T
A
C
G
T
A
C
T
A
C
G
T
A
G
T
Y
A
C
G
T
A
C
G
T
X
A
C
G
T
A
G
T
A
C
G
T
A
C
G
T
C
G
T
C
G
T
C
G
T
C
G
T
C
G
T
C
G
T
C
G
T
C
G
T
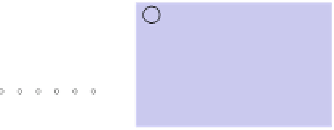
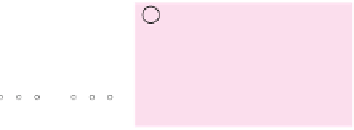
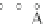
Fig. 5 The phylogeny-aware algorithm can be sensitive to errors in the guide phylogeny. The tree on the left
represents the true evolutionary history and the trees in the middle and on the right indicate the right and a
wrong order of aligning the sequences. The greedy calling of insertions (
PRANK
+F
) marks flagged gaps that are
re-used (curved right arrow) as permanent insertions. When the alignment order is correct (top), the algorithm
works perfectly. If A and C are incorrectly aligned first (bottom), the subsequent alignment of B appears to
confirm an insertion in C although the true event is a deletion shared by A and B. As the insertion in column 4
is marked permanent (filled square), the site belonging to that columns has to placed in a columns of its own.
The resulting alignment is too long and gappy. See Fig.
3
for the notation
a deletion may appear as an insertion and, by marking sites
incorrectly as a permanent insertion, the algorithm has to place
characters truly homologous to that to separate columns (Fig.
5
).
Although the resulting alignment is too long and gappy, small
errors in the alignment order may not be too serious in typical
evolutionary analyses: an incorrect alignment such as that in
Fig.
5
does not indicate all true homologies but neither does it
contain false homology statements.
In addition to the wrong alignment order, missing data can
cause errors with the
PRANK
+F
variant. The algorithm assumes that
alignment gaps are caused by insertions and deletions and then
chooses the most plausible explanation of the two. One isolated
gap caused by missing data may not be serious but if several
sequences lack data at the same region, the gap pattern created
may look like an insertion in the complete sequences; when this
region is falsely marked as a permanent insertion, the subsequent
alignment must place the affected region in separate columns. As
sequences are often truncated at their ends, the marking of terminal
gaps as permanent insertions is by default disabled by
PRANK
.