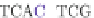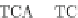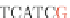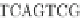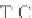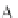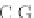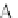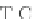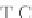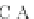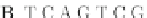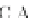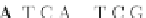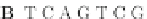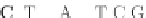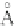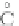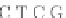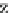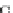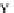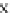Biology Reference
In-Depth Information
Fig. 4 The phylogeny-aware algorithm can distinguish and correctly align near-by insertion and deletions. The
tree on the left represents the evolutionary history of five short sequences undergoing two insertion and two
deletion events. The tree in the middle indicates how the alignment is divided into pairwise alignments of
sequences (or sequence graphs). The resulting alignments are shown on the right. The classical alignment
algorithm considers length differences as deletions and cannot place independent insertions into separate
columns; often it also moves near-by gaps and indicates false homologies, here resulting in substitutions.
A variant of the phylogeny-aware algorithm with greedy calling of insertions, known as
PRANK
+F
, considers the
re-use of a flagged gap as evidence that the gap was created by an insertion. It then changes the flags
indicating a pre-existing gap (filled diamond) to ones indicating a permanent insertion (filled square) and does
not allow matching of these sites at later alignments. This forces the correct placement of independent
insertions into separate alignment columns. The same functionality can be obtained with sequence graphs
and greedy pruning of the graph edges. See Fig.
3
for the notation
gap is re-used as permanent insertions that cannot be matched at
the later stages of the progressive alignment; to prevent overlapping
deletions from confirming embedded insertions, the re-use of a gap
has to be done for its full length with matching characters at the
flanking sites. This approach can separate multiple insertions at the
same position to independent events without effect on the place-
ment of gaps caused by deletions (Fig.
4
). When the order of
aligning the sequences is correct and the sequence sampling is
dense enough to call near-by gaps as separate events,
PRANK
+F