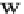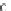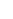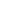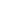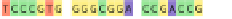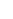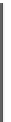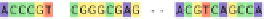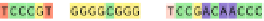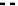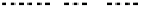Biology Reference
In-Depth Information
Fig. 1 The gap patterns in a true alignment reflect the underlying phylogeny of the sequences. Insertions and
deletions create gap patterns (boxes in the alignment; numbered at the bottom) that reflect the phylogenetic
locations of the events (black and gray bubbles in the tree). If multiple parallel insertions occur at homologous
positions (events 5, 6 and 7), inserted columns can be placed in any order without effect on the homology
statement. In the case of more than two parallel insertions, some inserted fragments are disconnected from
the rest of the alignment (here, event 6). The phylogenetic location and type of event 2 is uncertain and it could
also be a deletion in the sister branch. Whereas the phylogeny-aware algorithm would re-align the sequences
correctly, the classical progressive algorithm (here ClustalW [
7
]) fails to resolve the true insertion and deletion
events
certain site can be substituted several times, subsequent substitutions
may turn the character state back to an earlier one and characters in
different evolutionary lineages may independently obtain the same
new character state, all still remaining homologous to each other; (2)
an insertion adds new characters to the sequence and subsequent
insertions may be nested inside a fragment inserted by a preceding
insertion event but, as mentioned above, insertions in different
lineages are never homologous and evolve independently; and (3) a
deletion removes characters permanently and the characters once
deleted cannot be reverted, a potential insertion at the same position
introducing new characters that are not homologous with the
deleted ones.
By a comparison of two homologous sequences we can detect
sites that have undergone substitutions but, without more infor-
mation, we cannot tell which sequence has changed at which posi-
tion. In contrast to this, length differences between two sequences
can be explained by deletions of existing characters in one sequence
or insertions of new characters in the other. The evolutionary
lineage on which the substitution-differences between two
sequences have occurred can be resolved using outgroup sequences
and by inferring the character states for the ancestor of the two.