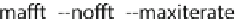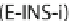Biology Reference
In-Depth Information
The number after
--maxiterate
specifies the number of cycles.
In the current version, even if a number larger than 16 is specified,
the calculation is finished at the 16th cycle. This is because major
improvements in alignment quality are made in early cycles.
An option, NW-NS-i, without the FFT algorithm,
is also
available.
In order to further improve accuracy, MAFFT partly uses the
consistency criterion. Several different types of consistency criteria
were described previously [
22
-
26
]. TCoffee [
25
] achieved a great
improvement in accuracy. ProbCons [
26
] and other methods
[
27
-
29
] are largely based on this idea. These methods generally
require long computational time. Unlike them, three iterative
refinement options of MAFFT, L-INS-i, G-INS-i, and E-INS-i
use a consistency criterion similar to COFFEE [
24
], combining it
with the WSP objective function, and maximize it using the itera-
tive refinement process.
2.3 Consistency
Criteria
These three options are designed for different types of input
sequences. G-INS-i is suitable forsequences that have homology
over the entire region, whereas L-INS-i and E-INS-iare suitable
for sequences that have homology only in partial regions.
See
[
30
]
formore detailed information.
2.4 RNA Alignments
The X-INS-i and Q-INS-i options are specially designed for RNA
alignment, considering secondary structures [
31
]. In Q-INS-i, the
base pairing probability is calculated by McCaskill's algorithm [
32
],
incorporated into the objective function, and an iterative refine-
ment method is applied to maximize the objective function. In
X-INS-i, secondary structure is also considered in the pairwise
comparison stage, by using SCARNA [
33
], in addition to the
objective function.
Both Q-INS-i and X-INS-i use source codes from the Vienna RNA
package [
34
], MXSCARNA [
35
] and ProbconsRNA [
26
]. Some
MAFFT packages do not contain these source codes and thus do
not support these options.