Biology Reference
In-Depth Information
Bayesian Information Criterion
50
0
−50
−100
0
1
2
3
4
5
Penalty level
Fig. 3.6
Graphical output of the
simone
package for dynamic Bayesian network learning via BIC
criterion minimization
257710_at 255764_at 255070_at 253425_at
0.0000000 0.0000000 0.0000000 0.0000000
253174_at 251324_at 245319_at 245094_at
0.0000000 0.0000000 0.0000000 -0.6961228
In addition to the LASSO, the
lars
package implements Least Angle Regression
(LAR) from
Efron et al.
(
2004
) and stepwise regression. Both can be fitted with the
same functions used for the LASSO by setting the
type
arguments of
lars
and
cv.lars
to either
"lar"
or
"stepwise"
, as shown below.
> lar.fit = lars(y = y, x = x, type = "lar")
> plot(lar.fit)
> lar.cv = cv.lars(y = y, x = x, type = "lar")
> step.fit = lars(y = y, x = x, type = "stepwise")
> plot(step.fit)
> step.cv = cv.lars(y = y, x = x, type = "stepwise")
The
simone
package for Statistical Inference for MOdular NEtworks by
Chiquet et al.
(
2009
) provides an implementation of the LASSO specifically tar-
geted to dynamic Bayesian network learning. Model estimation is performed using
the
simone
function with
clustering = FALSE
.
> library(simone)
> simone(arth12, type = "time-course")
The
simone
function allows clustering assumption, i.e., modular network. Model
estimation is now performed using the
simone
function with clustering.
> ctrl = setOptions(clusters.crit = "BIC")
> simone(arth12, type = "time-course",
+ clustering = TRUE, control = ctrl)
The optimal value of the
L
1
penalty can be chosen by minimizing the BIC cri-
terion, which is computed by
simone
when the model is fitted with
output =
"BIC"
.























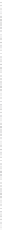







Search WWH ::

Custom Search