Biology Reference
In-Depth Information
1.0
0.8
0.6
0.4
0.2
significant arc
s
0.0
0.0
0.2
0.4
0.6
0.8
1.0
arc strengths
Fig. 2.9
Cumulative distribution function of the arc strength values computed with bootstrap re-
sampling from
dsachs
.The
vertical dashed lines
correspond to the estimated (
black
) and ad hoc
(
grey
) significance thresholds
The reason for the insensitivity of the averaged network to the value of the thresh-
old is apparent from the plot of
F
p
(
·
)
in Fig.
2.9
: arcs which are well supported by
the data are clearly separated from the ones that are not. Since the lowest strength
coefficient in the first set is 0
347 (i.e.,
the estimated threshold), any threshold that falls between those two values results in
thesameaveragednetwork.
.
962 and the highest one in the second set is 0
.
2.5.3 Handling Interventional Data
Usually, all the observations in a sample are collected under the same general con-
ditions. This is true both for observational data, in which treatment allocation is out-
side the control of the investigator, and for experimental data, which are collected
from randomized controlled trials. As a result, the sample can be modeled with a
single Bayesian network, because all the observations follow the same probability
distribution.
However, this is not the case when several samples resulting from different exper-
iments are analyzed together with a single, encompassing model. Such an approach
is called
meta-analysis
(see
Kulinskaya et al.
,
2008
, for a gentle introduction). First,
environmental conditions and other exogenous factors may differ between those
experiments. Furthermore, the experiments may be different in themselves; for ex-
ample, they may explore different treatment regimes or target different populations.
This is the case with the protein signaling data analyzed in
Sachs et al.
(
2005
). In
addition to the data set we have analyzed so far, which is subject only to a general


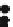
































Search WWH ::

Custom Search