Biology Reference
In-Depth Information
P
(
pakts473
)
P
(
PKA
)
without evidence
with evidence
without evidence
with evidence
HIGH
HIGH
AVG
AVG
LOW
LOW
0.0
0.2
0.4
0.6
0.2
0.4
0.6
probability
probability
Fig. 4.1
Probability distributions of
pakts473
and
PKA
before and after inhibiting
p44.42
> querygrain(jprop, nodes = "PKA")$PKA
PKA
LOW AVG HIGH
0.48908954 0.45116629 0.05974417
All the queries illustrated above can be easily changed to maximum a posteriori
queries by finding the largest element in the distribution of the target node.
> names(which.max(querygrain(jprop,
+
nodes = c("PKA"))$PKA))
[1] "LOW"
4.2.2 Approximate Inference
The conditional probability queries from the previous section produce similar
results when they are performed with the logic sampling algorithm, illustrated
in Algorithm
4.2
and implemented in
bnlearn
in the
cpdist
and
cpquery
functions.
> particles = cpdist(fitted, nodes = "pakts473",
+ evidence = (p44.42 == "LOW"))
> prop.table(table(particles))
particles
LOW AVG HIGH
0.665622103 0.332827458 0.001550438
> particles = cpdist(fitted, nodes = "PKA",
+
evidence = (p44.42 == "LOW"))
> prop.table(table(particles))
particles
LOW AVG HIGH
0.48865276 0.45108553 0.06026171























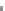







Search WWH ::

Custom Search