Environmental Engineering Reference
In-Depth Information
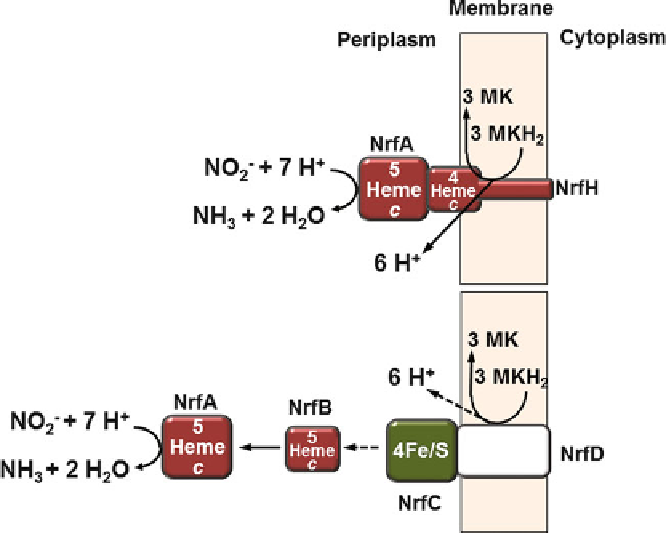
Figure 9 Electron transport chain models catalyzing the oxidation of menaquinol by nitrite in
representative respiratory Nrf systems. Top: Nrf system of
Wolinella succinogenes
. Bottom: Nrf
system of
Escherichia coli
. See text and Table
1
for details. The dashed arrows denote that proton
release to the periplasmic side of the membrane by NrfD as well as direct electron transfer between
NrfC and NrfB are speculative. For simplicity, only monomeric enzyme forms are shown. MK,
menaquinone; MKH
2
, menaquinol; Fe/S, iron-sulfur center. Modified from [
24
].
(NrfHA
2
)
2
complex, it is inferred that menaquinol binds at the periplasmic side of
the membrane in the vicinity of heme 1. Therefore, it is conceivable that protons are
released to the periplasmic space upon menaquinol oxidation. This would make the
catalysis of menaquinol oxidation by nitrite an electroneutral, i.e., non proton
motive force-generating, process [
24
,
73
,
77
].
Enteric bacteria such as
E. coli
do not encode a NrfH homolog in their
nrf
gene
clusters. Instead,
E. coli
was reported to employ a protein assembly consisting of
the proteins NrfB, NrfC, and NrfD in order to transfer electrons from menaquinol to
NrfA, which appears to be a soluble protein present in the periplasmic space
(Figure
9
, bottom and Table
1
, enzyme class 2.4). Mainly concluded from genetic
studies, a membrane-bound NrfCD complex was postulated to oxidize menaquinol
near the periplasmic side of the membrane [
78
-
80
]. This hypothesis is in line with
the experimentally proven location of a quinone binding site in the structurally
similar PsrCB subcomplex of a potential polysulfide reductase from
Thermus
thermophilus
[
81
]. As for the NrfHA complex, such a topology of the reactive
sites for menaquinol and nitrite in the NrfABCD system would make electrogenic
Search WWH ::

Custom Search