Biology Reference
In-Depth Information
a
b
Planar wave
Focal excitation
Spiral wave
Spiral Breakup
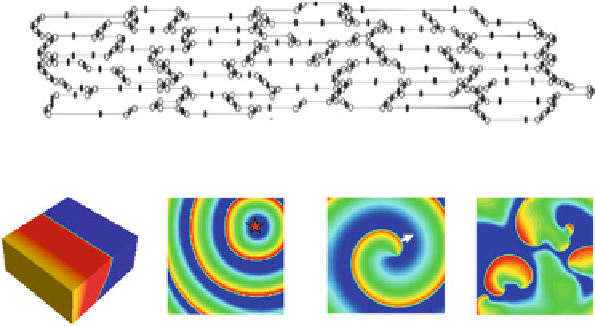
Fig. 10.6 (a) A schematic plot of a cell network reconstructed from real tissue (Spach and
Heidlage
1995
). (b) Excitation dynamics in cardiac tissue for normal rhythms (planar wave) and
arrhythmias (focal excitation, spiral reentry, and spiral wave breakup).
Arrows
indicate directions
of conduction. Voltage changes from low to high as the color changes from
blue
to
red
1962 following the Hodgkin-Huxley model (Noble
1962
), a great number of
advanced models have been developed and used to study cardiac excitation and
contraction dynamics in single cell, tissue, and whole heart models over the last
four decades, setting the heart to be the most well-modeled organ. Moreover, many
modeling studies are closely combined with experiments, which further enhance
the applicability of these models.
With the experimental and computational studies, our current understanding of
the mechanisms of cardiac arrhythmias has been greatly improved, however,
effective therapeutics that can prevent cardiac arrhythmias are still lacking. Most
of the antiarrhythmic drugs that were developed based on certain mechanistic
insights of arrhythmias are not effective but rather cause more mortality (CAST
1989
; Waldo et al.
1996
). One can easily argue that we still do not understand the
mechanisms due to the multi-scale complex regulations of the excitation and
contraction dynamics in the heart. A full understanding of the system would be
required to investigate the dynamics at each scale and how the dynamics at one
scale affects the dynamics at another scale using both experimental and computa-
tional approaches. Therefore, systems biology approaches that combine computa-
tional biology and experimental biology are the likely means.
However, there are many challenges ahead. On the experimental side, one of the
major problems is how to obtain the molecular and cellular information accurately
that can be faithfully used in modeling. On the computational side, since one cannot
develop a model of a heart or even a piece of tissue at the scale of molecules due to
computational limitations and complexity, a multi-scale modeling strategy is
needed. However, how to develop such a strategy and maintain the fidelity of
information when changing from one scale to another of modeling is a big chal-
lenge (Qu et al.
2011
). One possible solution is to closely combine experiments and

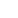
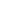

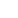






Search WWH ::

Custom Search