Biology Reference
In-Depth Information
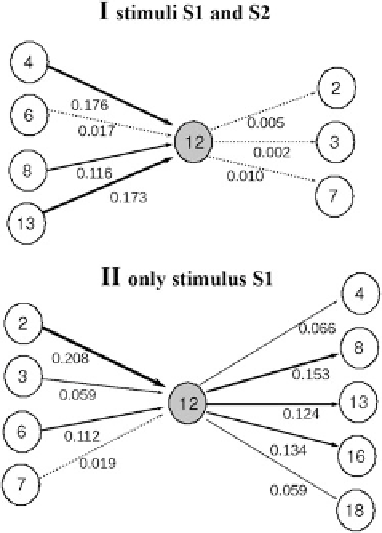
Fig. 8.6
The dynamical structure of biomolecular information flows modify the catalytic activity
of self-organized multienzymatic complexes
. The figure displays the in and out biomolecular
information flows belonging to the metabolic core (
gray circle
). The white circles represent active
multienzymatic complexes. The metabolic core (the metabolic subsystem 18) depends on the
biomolecular information contained in substrate fluxes and regulatory signals; this core performs
three functions simultaneously: signal reception and integration, and being a source for new
biomolecular information. The dynamical functional structure depends strongly on the external
stimuli (I and II)
plasticity. These drastic changes are self-regulated by means of Module
α
,an
emergent Module
, but now in this module all the directions of
effective connectivity reverse. Therefore, the transitions between the modules
provoke permanent changes in all catalytic activities of the self-organized enzy-
matic subsystems, and these metabolic switches are triggered by changes in the
external conditions.
Functionally, metabolic switches are discrete transitions between catalytic
modules regulated by systemic dynamics of the biomolecular information flows.
At a molecular level, a switch is a biochemical process in which determined
enzymes can undergo a persistent change in their catalytic status. Experimentally,
it has been observed that the molecular mechanisms involved in a switch depend on
posttranslational modification of enzymatic activities (Miller et al.
2005
) such as
phosphorylation (Oesch-Bartlomowicz and Oesch
2003
), acetylation (Pasini
et al.
2010
), and methylation (He et al.
2005
). Molecular switches have been
implicated in several metabolic processes including transcriptional regulation
δ
, and Module
β
Search WWH ::

Custom Search