Biology Reference
In-Depth Information
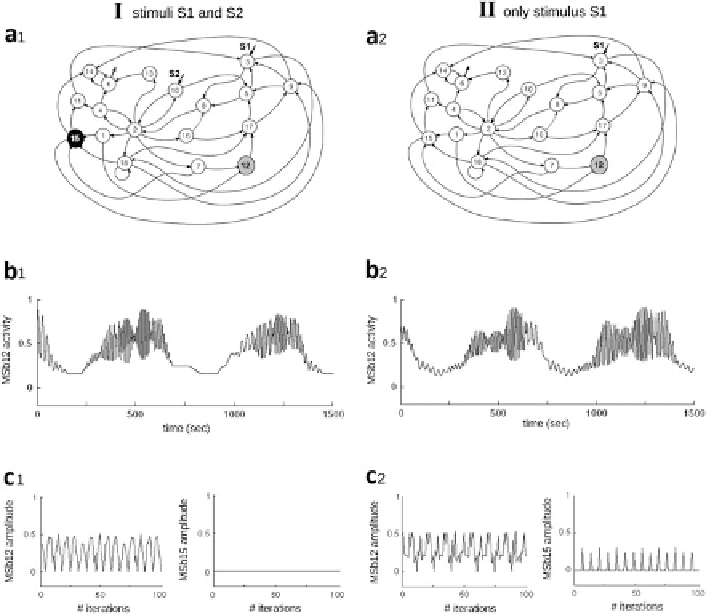
Fig. 8.4
Dynamic catalytic behaviors of the dissipative metabolic network
. The network
consisting of 18 self-organized multienzymatic complexes (metabolic subsystems) is depicted.
The interconnection by fluxes through the network, and substrate input fluxes are shown. For
simplification purposes, the topological architecture of regulatory signals considered in the
network is not indicated. The network was perturbed by two different external conditions:
(I) two substrate input fluxes S1 and S2 (
left column in panel
) and (II) only with the stationary
stimulus S1 (
right column
)(
Panel
a1). Under condition I, a systemic metabolic structure sponta-
neously emerges in the network in which the enzymatic subsystem MSb12 is always active
(metabolic core) (
gray circle
) and the catalytic subsystem MSb15 is inactive (
black circle
),
whereas the rest of enzymatic sets exhibit on-off changing states (
white circle
)(
Panel
a1).
Under condition II, the network preserves the systemic metabolic structure exhibiting flux
plasticity which involves persistent changes in all the catalytic patterns (
Panels
b and c) and
structural plasticity resulting in a persistent change in the dynamic state of the subsystem MSb15
which exhibits a transition from an off (
black circle
) to an on-off changing state (
white circle
)
(
Panel
a2). Enzymatic activities of the MSb12 (metabolic core) exhibit a large number of different
catalytic transitions between periodic oscillations and some steady states (
Panel
b1). Condition II
produces persistent changes in the catalytic activity of subsystem MSb12 without steady states
(flux plasticity) (
Panel
b2). Self-regulation the network under external conditions I and II implies
changes in the amplitude of the activities from metabolic subsystem activities (flux plasticity)
(
Panels
c1, c2). Adapted from Fig. 2 in (De la Fuente et al.
2011
)
Search WWH ::

Custom Search