Biomedical Engineering Reference
In-Depth Information
10.1
Introduction
A variety of epigenetic modifi cations modulate eukaryotic genomic information
affecting transcription profi les and DNA replication and repair processes. Many
investigators in the fi eld of epigenomics focus on DNA methylation and histone
modifi cations and how these modulations interact to form stable epigenetic chroma-
tin conformation. In this review, we focus on the methods used to assay DNA
methylation.
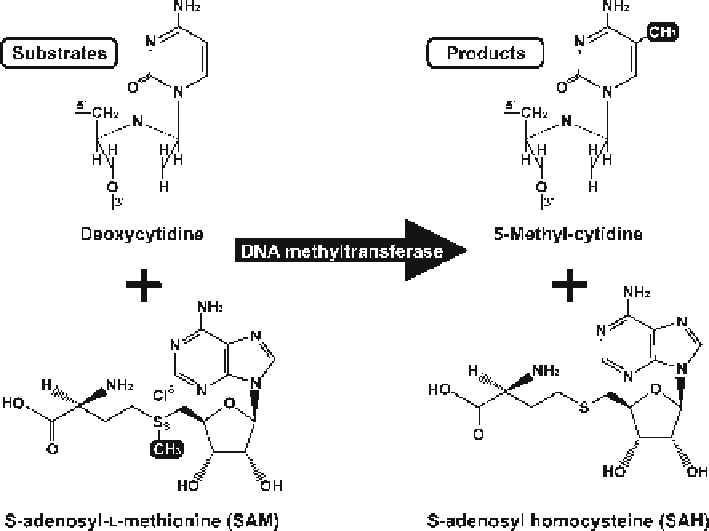
DNA methylation involves the addition of a methyl moiety to the cytosine-5
position (Fig.
10.1
) in most eukaryotic organisms. The enzymes that specifi cally
methylate the C5 carbon of cytosines in DNA to produce C5-methylcytosine are
DNA methyltransferases (DNMTs). Enzymatically, they transfer a methyl group
which is derived from
S
-adenosyl- L-methionine to the C5 carbon of the pyrimidine
ring. This results in the generation of 5-methyl-cytosine and
S -adenosyl-
homocysteine
. Five different enzymes involved in DNA methyl-group transfer are
known: DNMT1 maintains DNA methylation after replication by methylating the
newly synthesized strand (Araujo et al.
1999
), DNMT3A and DNMT3B methylate
genomic regions de novo (Takeshima et al.
2006
), DNMT2 has a tRNA methylating
Fig. 10.1
Chemical reaction for methylation on cytosine residues