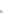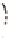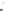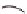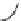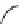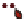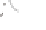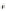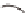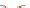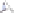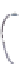Biomedical Engineering Reference
In-Depth Information
a
b
Parental strains
Enhancer or Promoter
GAL4 gene
X
JO-GAL4
UAS-cameleon 2.1
GAL4
“Driver strain”
“Responder strain”
+
50 µm
Progeny
GECI
UAS
c
Cameleon 2.1
JO-GAL4 / UAS-cameleon 2.1
Tissue-specific
enhancer
Exogeneous
transcription factor
(e.g. GAL4 gene)
Endogeneous gene that is expressed in
a tissue-specific manner
Fig. 7.1
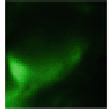
Genetic tools to express a GECI construct in tissue-specifi c manner. (
a
) Illustration of the
binary GAL4/UAS expression system. In the transgenic progeny obtained from a genetic cross of
a GAL4 strain and a UAS-GECI strain, the indicator is expressed in a specifi c cell population
defi ned by the parental GAL4 strain (Brand and Perrimon
1993
). (
b
) A genetic cross to obtain
Cameleon 2.1 expression in Johnston's organ (JO) neurons. JO-GAL4 is a driver strain that
expresses GAL4 in JO neurons (Kamikouchi et al.
2009
). When this GAL4 fl y is crossed to the
responder strain UAS-Cameleon 2.1 (Fiala and Spall
2003
), progeny containing both elements is
produced. The presence of GAL4 drives expression of Cameleon 2.1 (
green
) in the JO neurons.
The
dotted line
indicates the array of cell bodies of JO neurons. (
c
) The enhancer-trapping method.
A
P
element carrying an exogenous transcription factor such as the GAL4 transcriptional activator
is randomly mobilized throughout the genome, bringing the expression of GAL4 under the control
of endogenous tissue-specifi c enhancers (Duffy
2002
)
enables a transgene expression in clones of neurons derived from a common ancestor
cell (Kohatsu et al.
2011
). Various so-called intersectional strategies have also been
developed to refi ne the pattern of GAL4-driven gene expression, such as the expres-
sion of the GAL4 inhibitor GAL80 in addition to GAL4 but in a different, overlap-
ping subset of cells (Lee and Luo
1999
), or the random induction of transgene
expression using a fl ippase (Flp) and other recombinases (Nern et al.
2011
).
In the “split GAL4” system, a functional GAL4 protein can also be created by an
independent expression of two parts of it, and the expression of both parts can be
genetically targeted to different, overlapping subsets of cells (Luan et al.
2006
).
Lastly, alternative binary systems have recently been described, such as the LexA