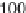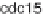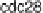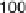Graphics Reference
In-Depth Information
data sets to infer the regulation of latent factors (to modelprotein, RNA degradation
and other factors that cannot be measured by microarrays) on genes by structure
equation modeling.
Without augmenting data in the aforementioned four data sets, those time points
(sample sizes) are not su
cient to obtain statistical inferences. However, in order to
carefully distinguish which data sets among alpha, cdc
, cdc
and Elu to integrate,
Shieh et al. (
) applied the algorithm in Bar-Joseph et al. (
) to align the four
data sets in the same time domain. Figure
.
clearly shows that alpha, cdc
and
cdc
exhibit similar trends, while Elu does not. his observation is consistent with
the following biological argument. he Elu data set was synchronized by elutriation,
unlike the other three sets, which were treated by pheromone α factor, cdc
and
cdc
mutant, respectively.BasedonFig.
.
,theyintegrated thosethreedatasetsand
applied the proposed structural equation modeling algorithm to the integrated data
in order to infer genetic interactions. A gene network of five genes that are synthetic
Figure
.
.
he expression levels of two genes in the four aligned data sets; each gene's expression
pattern in Elu is clearly different from its corresponding patterns in the other sets