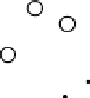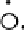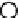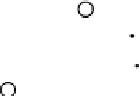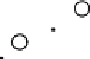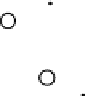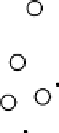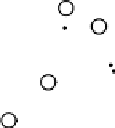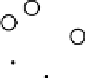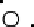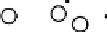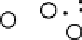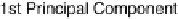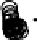Graphics Reference
In-Depth Information
Example 6
In this example we use the “Pima diabetes” data set from the UCI Ma-
chine Learning data archives. he reduced kernel method is adopted by randomly
sampling
% of the column vectors from the full kernel matrix. For reduced kernel
PCA, we assume K is the underlying reduced kernel data matrix of size n
6
m,where
n is the data size and m is the reduced set size (i.e.,column set size). KPCA using the
reduced kernel is a singular value decomposition problem that involves extracting
the leading right and let singular vectors β and β:
′
n
n
K β
αβ,normalizedtoα β
′
β
′
I
n
andαβ
β
.
(
.
)
−
=
=
=
n
Inthis data set, there arenine variables, including number of times pregnant, plasma
glucoseconcentration (glucosetolerancetest),diastolicbloodpressure(mmHg),tri-
cepsskin foldthickness (mm),two-hour seruminsulin (mu U/ml),body massindex
(weight in kg/(height in m)
), diabetes pedigree function, age (years), and the class
variable (testfordiabetes),whichcanbepositive ornegative. Fordemonstration pur-
poses, we use the first eight variables as input measurements and the last variable as
Figure
.
.
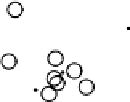
Results from PCA based on original input variables
Figure
.
.
Results from KPCA with the polynomial kernel of degree
and scale