Biology Reference
In-Depth Information
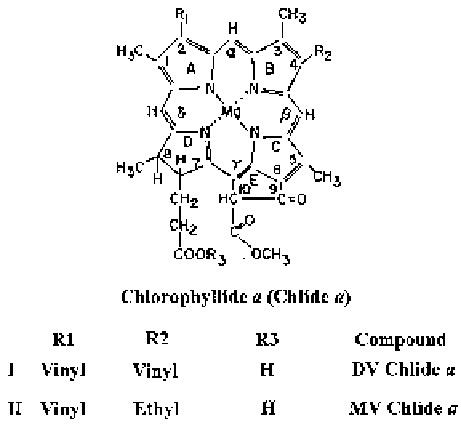
Fig. 9.1 DV and MV
chlorophyllide (Chlide)
a
9.1 Formation of Chlide
a
via Light-Independent
Pchlide
a
Reductase(s)
Algae, ferns, mosses, and the cotyledons of most gymnosperms, all with a
DDV-LDV-LDDV greening group affiliation (Ioannides et al.
1994
) are capable
of converting Pchlide
a
to Chlide
a
in the absence of light (Kirk and Tilney-Basset
1967
; Rudiger and Schoch
1991
; Shulz and Senger
1993
), via a reaction catalyzed
by a light-independent Pchlide
a
reductase. Most probably light-independent
Chlide
a
formation is via a nt-SW-Pchlide a species (Schoefs and Franck
1998
).
Although in angiosperms light is required for the formation of photosynthetic
pigment-protein complexes and the accumulation of massive amounts of Chl,
Adamson and coworkers pioneered the notion that in this phylum, a certain amount
of Chl
a
biosynthesis can also take place in darkness via a light-independent Pchlide
a
reductase (Adamson et al.
1997
). Dark-incorporation of
14
C-glutamate and
14
C-
ALA into
14
C-Chl
a
in barley leaves and barley etiochloroplasts appear to confirm
Adamson's contention (Tripathy and Rebeiz
1987
). It is therefore proposed that
angiosperms such as cucumber and barley. It should be emphasized however that
the amount of Chl
a
formed via Chlide
a
in darkness is very small, and its biological
significance is unknown.
Genetic and sequence analysis have indicated that in
R. capsulatus
, three genes,
bchL
,
bchN
,and
bchB
appear to be involved in Pchlide
a
reduction in darkness
(Suzuki et al.
1997
). The three open frames exhibited significant sequence similarity
to the three subunits of nitrogenase, which led to the proposal that light-independent
Pchlide
a
reductase and nitrogenase share a common evolutionary ancestor. Expres-
sion of the
bchL
,
bchN
,and
bchB
genes has been however unsuccessful. Very
Search WWH ::

Custom Search